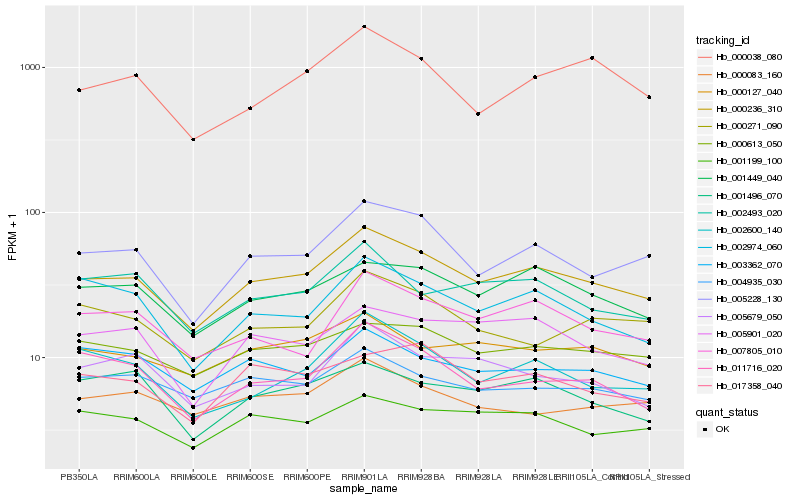
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_310 |
0.0 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 2 |
Hb_005228_130 |
0.0697017447 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_001449_040 |
0.0719123253 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000613_050 |
0.0809545949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011716_020 |
0.0850820458 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 6 |
Hb_000127_040 |
0.0857751737 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 7 |
Hb_002600_140 |
0.0859455736 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 8 |
Hb_001199_100 |
0.0866156672 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 9 |
Hb_002974_060 |
0.0867209492 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 10 |
Hb_007805_010 |
0.0878602219 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 11 |
Hb_005901_020 |
0.087860695 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_004935_030 |
0.0900470751 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 13 |
Hb_002493_020 |
0.0910116065 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 14 |
Hb_017358_040 |
0.0910668069 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 15 |
Hb_000038_080 |
0.0911701681 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 16 |
Hb_000083_160 |
0.0915730691 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 17 |
Hb_005679_050 |
0.0919639814 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001496_070 |
0.0935132288 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_000271_090 |
0.0939033928 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_003362_070 |
0.0939070648 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |