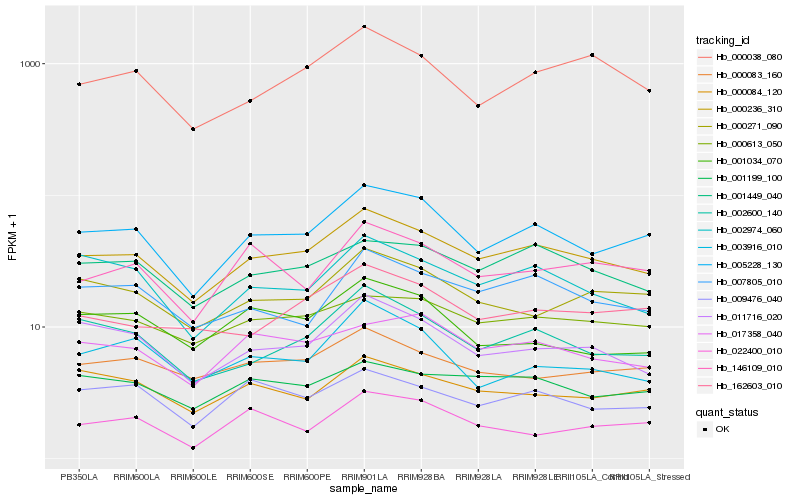
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005228_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_000236_310 |
0.0697017447 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 3 |
Hb_002600_140 |
0.091004704 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |
| 4 |
Hb_000271_090 |
0.1022605573 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_017358_040 |
0.102827787 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 6 |
Hb_009476_040 |
0.1037289515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011716_020 |
0.1047938296 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 8 |
Hb_000084_120 |
0.1051593202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g36240 [Jatropha curcas] |
| 9 |
Hb_002974_060 |
0.105992617 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X2 [Jatropha curcas] |
| 10 |
Hb_146109_010 |
0.1085268597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001449_040 |
0.1098553466 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007805_010 |
0.1100205815 |
- |
- |
PREDICTED: ribosome biogenesis protein BMS1 homolog [Jatropha curcas] |
| 13 |
Hb_000613_050 |
0.1104749231 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003916_010 |
0.1131183509 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 15 |
Hb_000083_160 |
0.1131376153 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 16 |
Hb_162603_010 |
0.113418344 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_001199_100 |
0.1156883018 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 18 |
Hb_001034_070 |
0.1176306026 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000038_080 |
0.1188171167 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 20 |
Hb_022400_010 |
0.1194957847 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |