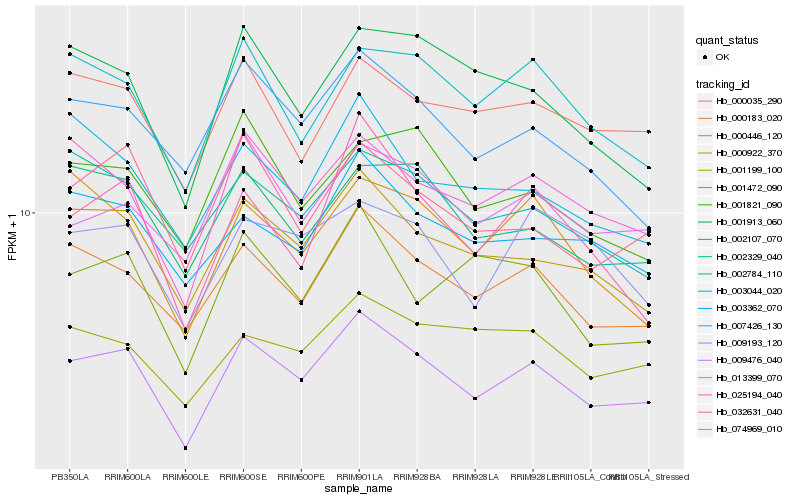
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009476_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000183_020 |
0.0641691138 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000922_370 |
0.0658830345 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 4 |
Hb_025194_040 |
0.0703027174 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 5 |
Hb_074969_010 |
0.0789061579 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 6 |
Hb_007426_130 |
0.0801732331 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 7 |
Hb_002784_110 |
0.0806045004 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 8 |
Hb_000035_290 |
0.0814669605 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 9 |
Hb_002107_070 |
0.0817342412 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 10 |
Hb_003362_070 |
0.0818229957 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 11 |
Hb_013399_070 |
0.0834697611 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 12 |
Hb_032631_040 |
0.0847465415 |
- |
- |
PREDICTED: uncharacterized protein LOC105638769 [Jatropha curcas] |
| 13 |
Hb_001199_100 |
0.0847866703 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 14 |
Hb_009193_120 |
0.0870664849 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 15 |
Hb_002329_040 |
0.087163282 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 16 |
Hb_003044_020 |
0.0883485161 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 17 |
Hb_000446_120 |
0.0887700889 |
- |
- |
PREDICTED: uncharacterized transmembrane protein DDB_G0289901 [Jatropha curcas] |
| 18 |
Hb_001913_060 |
0.0887929336 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 19 |
Hb_001821_090 |
0.0895200738 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 20 |
Hb_001472_090 |
0.0896549603 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |