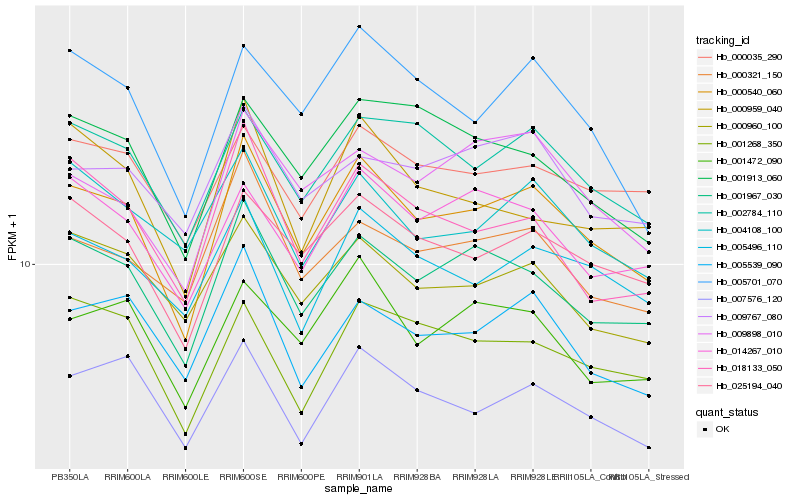
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 2 |
Hb_001967_030 |
0.0605421913 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 3 |
Hb_025194_040 |
0.0683033964 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002784_110 |
0.0772740344 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 5 |
Hb_007576_120 |
0.0792379808 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 6 |
Hb_005496_110 |
0.0817579012 |
- |
- |
PREDICTED: uncharacterized protein LOC105107584 [Populus euphratica] |
| 7 |
Hb_001472_090 |
0.0833399005 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005701_070 |
0.0836313775 |
- |
- |
PREDICTED: uncharacterized TPR repeat-containing protein At1g05150-like [Jatropha curcas] |
| 9 |
Hb_005539_090 |
0.0837396011 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014267_010 |
0.0851338208 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000960_100 |
0.0858695718 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 12 |
Hb_009898_010 |
0.085989736 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 13 |
Hb_000321_150 |
0.0867086155 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000035_290 |
0.0868968662 |
- |
- |
RNA recognition motif-containing family protein [Populus trichocarpa] |
| 15 |
Hb_018133_050 |
0.0879303463 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 16 |
Hb_001913_060 |
0.0887352728 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 17 |
Hb_001268_350 |
0.0888697904 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |
| 18 |
Hb_004108_100 |
0.0915617238 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 19 |
Hb_009767_080 |
0.0927360784 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 20 |
Hb_000959_040 |
0.0937245268 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |