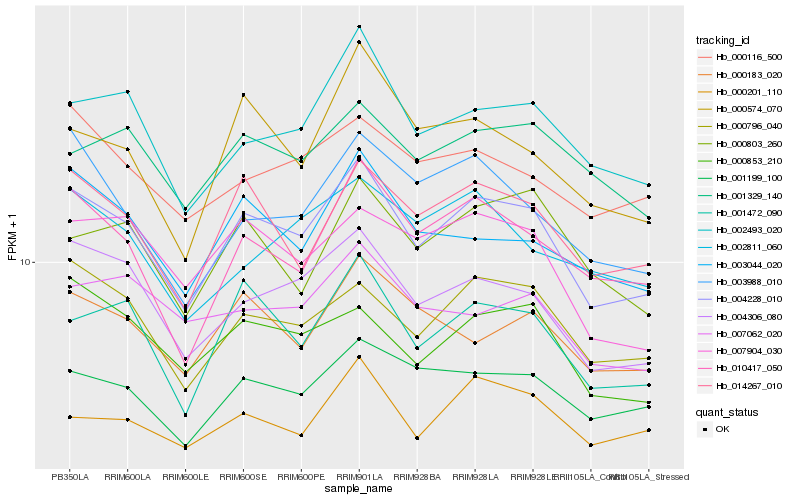
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004228_010 |
0.0 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |
| 2 |
Hb_004306_080 |
0.0615632465 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 3 |
Hb_000796_040 |
0.0664076096 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_014267_010 |
0.0665595995 |
- |
- |
PREDICTED: dnaJ homolog subfamily C GRV2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001472_090 |
0.0690172025 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010417_050 |
0.0711226961 |
- |
- |
PREDICTED: uncharacterized protein LOC105645746 [Jatropha curcas] |
| 7 |
Hb_002493_020 |
0.0741046518 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 8 |
Hb_001199_100 |
0.0762422034 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 9 |
Hb_000853_210 |
0.077184882 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003044_020 |
0.0785071448 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 11 |
Hb_007904_030 |
0.0809195162 |
- |
- |
PREDICTED: nuclear mitotic apparatus protein 1 [Jatropha curcas] |
| 12 |
Hb_000183_020 |
0.0833889499 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003988_010 |
0.0839082381 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |
| 14 |
Hb_000574_070 |
0.0857630305 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 15 |
Hb_000116_500 |
0.086129728 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 16 |
Hb_001329_140 |
0.0864794015 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 17 |
Hb_000201_110 |
0.086702879 |
- |
- |
hypothetical protein JCGZ_21690 [Jatropha curcas] |
| 18 |
Hb_000803_260 |
0.0876865016 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 19 |
Hb_002811_060 |
0.090013843 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 20 |
Hb_007062_020 |
0.091218537 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |