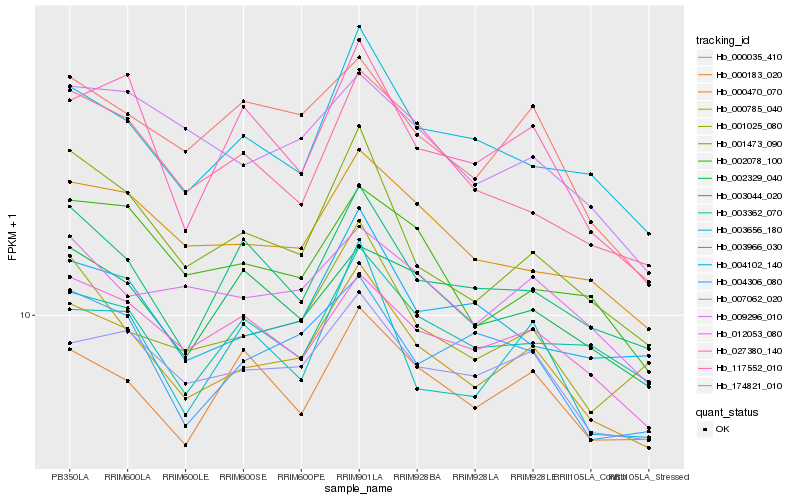
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000785_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 2 |
Hb_007062_020 |
0.0546959518 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 3 |
Hb_000470_070 |
0.0628159842 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 4 |
Hb_003966_030 |
0.0645415422 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 5 |
Hb_027380_140 |
0.0666325797 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001025_080 |
0.0672046366 |
- |
- |
hypothetical protein JCGZ_11431 [Jatropha curcas] |
| 7 |
Hb_001473_090 |
0.0687279097 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 8 |
Hb_000035_410 |
0.0690059313 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 9 |
Hb_004306_080 |
0.072227886 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 10 |
Hb_012053_080 |
0.0726642431 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 11 |
Hb_004102_140 |
0.0733164755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009296_010 |
0.0734544659 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 13 |
Hb_003044_020 |
0.0736385938 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 14 |
Hb_000183_020 |
0.0748012844 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002078_100 |
0.0749548125 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 16 |
Hb_002329_040 |
0.0750280132 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 17 |
Hb_174821_010 |
0.0751889661 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 18 |
Hb_003362_070 |
0.0752157115 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 19 |
Hb_117552_010 |
0.078075644 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003656_180 |
0.0797809513 |
- |
- |
PREDICTED: uncharacterized protein LOC105637927 [Jatropha curcas] |