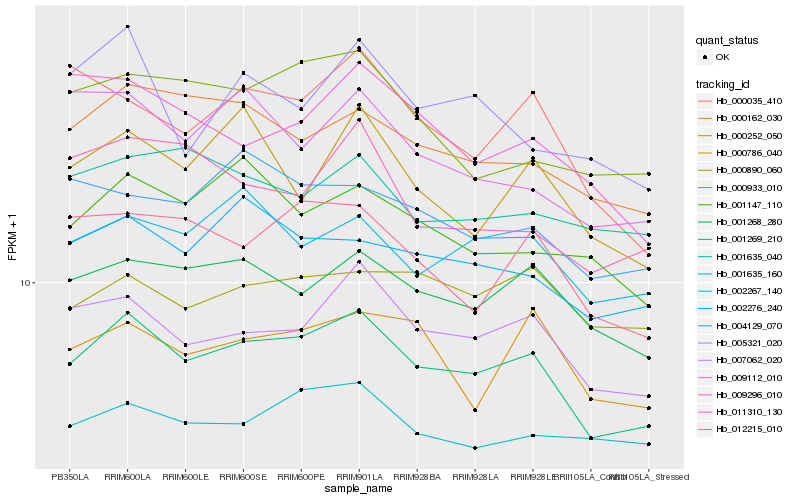
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_210 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007062_020 |
0.0626733823 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 3 |
Hb_001268_280 |
0.0718008509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000162_030 |
0.0797340643 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_002276_240 |
0.0813614939 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 6 |
Hb_012215_010 |
0.0818602559 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 7 |
Hb_002267_140 |
0.0825818928 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 8 |
Hb_000252_050 |
0.0872804139 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 9 |
Hb_001147_110 |
0.0887556409 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 10 |
Hb_000933_010 |
0.0893542685 |
- |
- |
- |
| 11 |
Hb_000035_410 |
0.0898337736 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 12 |
Hb_001635_160 |
0.0902947156 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 13 |
Hb_004129_070 |
0.0903036612 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 14 |
Hb_000890_060 |
0.0919906234 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 15 |
Hb_000786_040 |
0.0929149954 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 16 |
Hb_009296_010 |
0.0940614675 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 17 |
Hb_005321_020 |
0.0953833731 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 18 |
Hb_009112_010 |
0.0955716076 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 19 |
Hb_001635_040 |
0.0958180772 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_011310_130 |
0.0966419713 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |