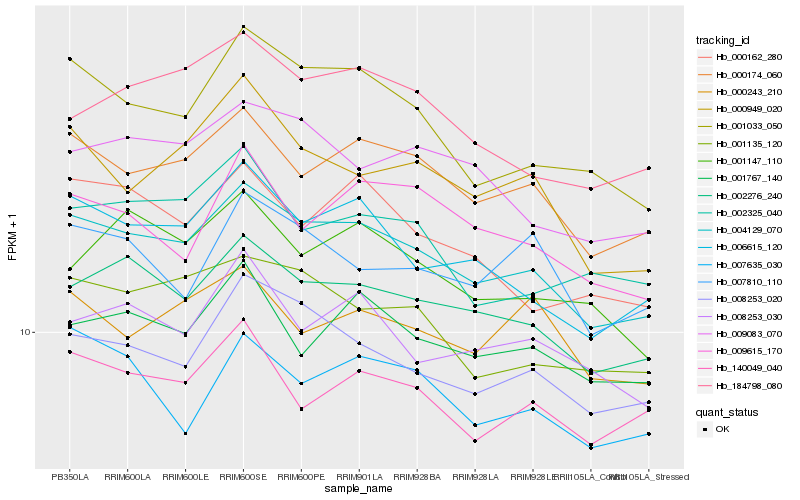
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_070 |
0.0 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 2 |
Hb_000174_060 |
0.0389276753 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_002276_240 |
0.0419357439 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 4 |
Hb_008253_020 |
0.0487806914 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 5 |
Hb_006615_120 |
0.0511945656 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 6 |
Hb_001033_050 |
0.0559581725 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 7 |
Hb_001135_120 |
0.0567859599 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001767_140 |
0.0609025764 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 9 |
Hb_009083_070 |
0.0613477885 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 10 |
Hb_140049_040 |
0.0613629454 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 11 |
Hb_008253_030 |
0.0628121461 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 12 |
Hb_000949_020 |
0.0654118404 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 13 |
Hb_184798_080 |
0.065684827 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 14 |
Hb_000162_280 |
0.0669359113 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 15 |
Hb_000243_210 |
0.0683204928 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_007635_030 |
0.0684866544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_009615_170 |
0.0687807258 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007810_110 |
0.0688161538 |
- |
- |
PREDICTED: DNA/RNA-binding protein KIN17 [Jatropha curcas] |
| 19 |
Hb_002325_040 |
0.0694533166 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001147_110 |
0.0696434973 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |