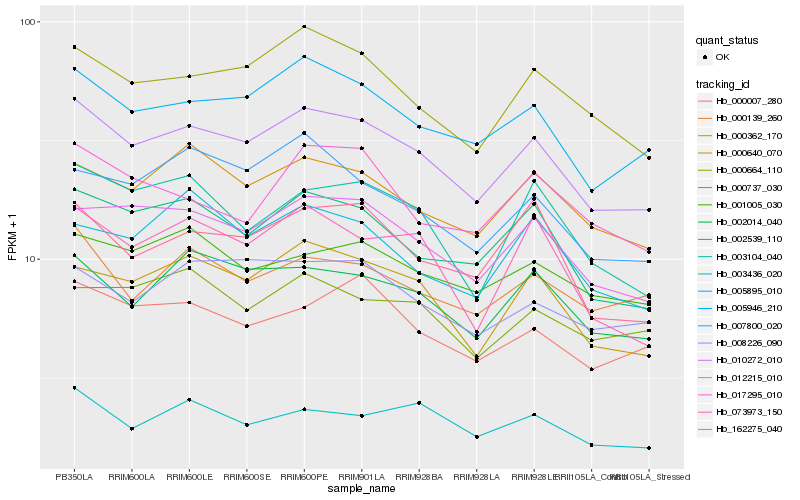
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010272_010 |
0.0 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 2 |
Hb_005946_210 |
0.0500131874 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 3 |
Hb_012215_010 |
0.054223013 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 4 |
Hb_002539_110 |
0.0581294822 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 5 |
Hb_000640_070 |
0.0591653206 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 6 |
Hb_000664_110 |
0.059523049 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 7 |
Hb_002014_040 |
0.060884774 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 8 |
Hb_000362_170 |
0.0631318589 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 9 |
Hb_000007_280 |
0.0718699847 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 10 |
Hb_162275_040 |
0.0721172671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001005_030 |
0.0736960305 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_008226_090 |
0.0739114645 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003436_020 |
0.07434909 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003104_040 |
0.0750830031 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 15 |
Hb_000139_260 |
0.0752293977 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 16 |
Hb_007800_020 |
0.0756718656 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_073973_150 |
0.0759748569 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000737_030 |
0.0800239488 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005895_010 |
0.081380677 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 20 |
Hb_017295_010 |
0.0820748954 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |