| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
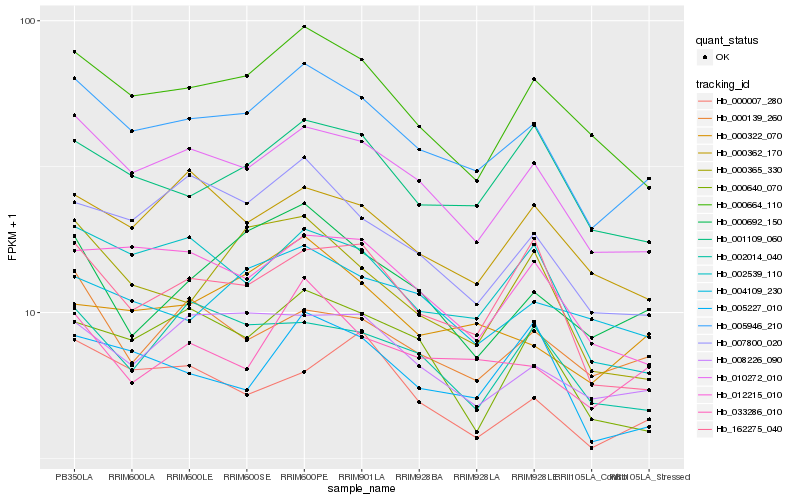
Hb_005946_210 |
0.0 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 2 |
Hb_010272_010 |
0.0500131874 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 3 |
Hb_000664_110 |
0.0691607285 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 4 |
Hb_001109_060 |
0.0758017644 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_008226_090 |
0.076073972 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012215_010 |
0.0762183643 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 7 |
Hb_002539_110 |
0.0767390594 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 8 |
Hb_005227_010 |
0.0777788132 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 9 |
Hb_033286_010 |
0.0784349477 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 10 |
Hb_007800_020 |
0.0800986808 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000322_070 |
0.0813211725 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_162275_040 |
0.0838420523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000640_070 |
0.084718607 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 14 |
Hb_000362_170 |
0.0854949905 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_004109_230 |
0.086014818 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000007_280 |
0.0862060828 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 17 |
Hb_000365_330 |
0.0864897762 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_000692_150 |
0.0865259819 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 3 [Jatropha curcas] |
| 19 |
Hb_002014_040 |
0.0868481426 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 20 |
Hb_000139_260 |
0.0870935719 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |