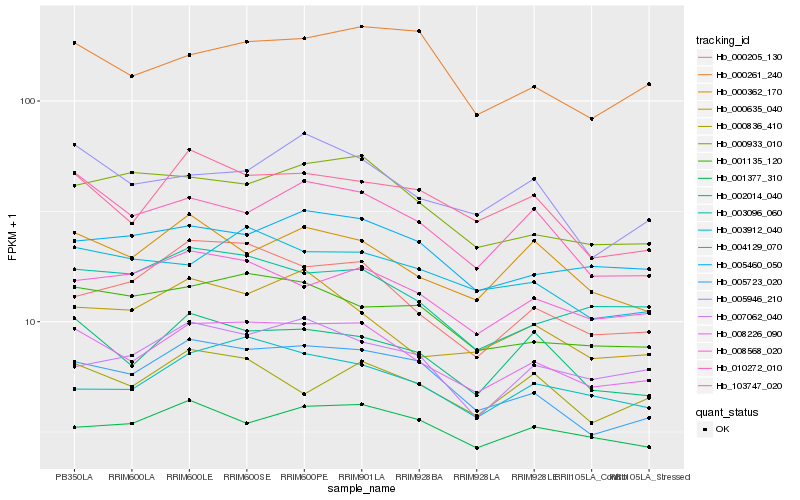
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008226_090 |
0.0 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005723_020 |
0.0642259995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002014_040 |
0.0649329433 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 4 |
Hb_008568_020 |
0.0661904374 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 5 |
Hb_001135_120 |
0.0674242662 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_103747_020 |
0.0679361118 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 7 |
Hb_003912_040 |
0.0682332864 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 8 |
Hb_005460_050 |
0.0688756411 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 9 |
Hb_003096_060 |
0.0691020506 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 10 |
Hb_004129_070 |
0.0702516815 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 11 |
Hb_000362_170 |
0.0729625495 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_010272_010 |
0.0739114645 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 13 |
Hb_000933_010 |
0.0751073345 |
- |
- |
- |
| 14 |
Hb_005946_210 |
0.076073972 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 15 |
Hb_000635_040 |
0.0763432699 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 16 |
Hb_000836_410 |
0.0764638438 |
- |
- |
sec10, putative [Ricinus communis] |
| 17 |
Hb_000205_130 |
0.0768643061 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_001377_310 |
0.0769467151 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 19 |
Hb_007062_040 |
0.0772876459 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 20 |
Hb_000261_240 |
0.0777178994 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |