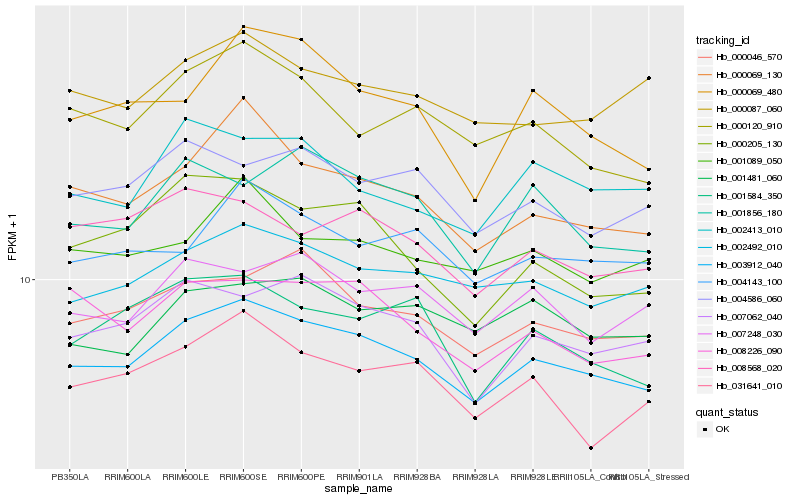
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003912_040 |
0.0 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 2 |
Hb_000069_130 |
0.0589202218 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.0605620312 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_002492_010 |
0.0625551137 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 5 |
Hb_007062_040 |
0.0631917949 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 6 |
Hb_008226_090 |
0.0682332864 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000120_910 |
0.068976643 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 8 |
Hb_000069_480 |
0.071385694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000205_130 |
0.0717552568 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_001856_180 |
0.0722556377 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_004143_100 |
0.07236651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007248_030 |
0.0734920808 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 13 |
Hb_004586_060 |
0.0735480549 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 14 |
Hb_001089_050 |
0.0739284745 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_002413_010 |
0.0747582746 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_008568_020 |
0.0748294493 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 17 |
Hb_031641_010 |
0.0754083826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001584_350 |
0.0754433639 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 19 |
Hb_000087_060 |
0.0755936663 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_001481_060 |
0.0757191893 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |