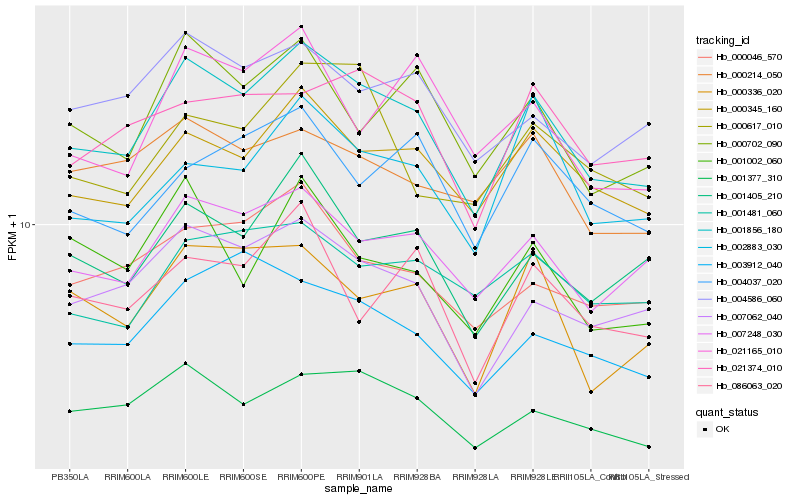
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_180 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 2 |
Hb_000345_160 |
0.0557662621 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 3 |
Hb_007248_030 |
0.0569103804 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 4 |
Hb_007062_040 |
0.0583712938 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 5 |
Hb_001405_210 |
0.0622076071 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 6 |
Hb_000702_090 |
0.0652768075 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_001377_310 |
0.0661849743 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 8 |
Hb_000046_570 |
0.0664133709 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 9 |
Hb_000336_020 |
0.0677926436 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 10 |
Hb_004586_060 |
0.0686079492 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 11 |
Hb_003912_040 |
0.0722556377 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 12 |
Hb_021374_010 |
0.0725805415 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_000214_050 |
0.0739715151 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001481_060 |
0.0750319455 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002883_030 |
0.0751874847 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 16 |
Hb_086063_020 |
0.0754658685 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 17 |
Hb_001002_060 |
0.076082046 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 18 |
Hb_004037_020 |
0.0768094578 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000617_010 |
0.0769375116 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 20 |
Hb_021165_010 |
0.0781676595 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |