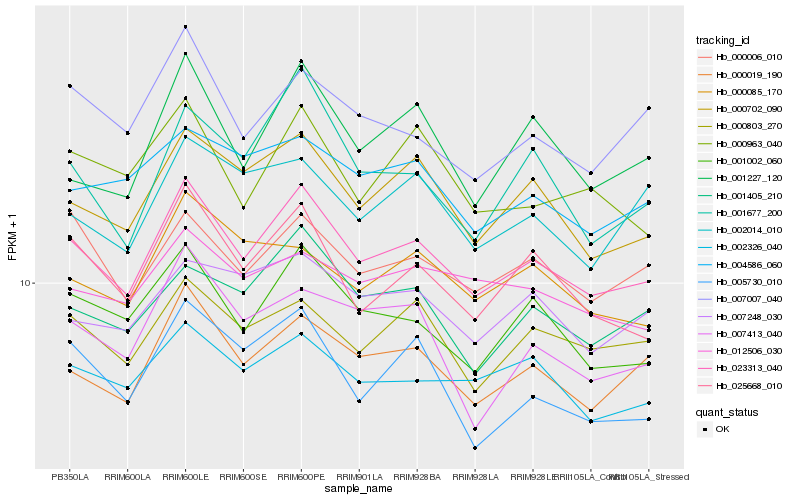
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023313_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000019_190 |
0.0516727286 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 3 |
Hb_000702_090 |
0.0567689487 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 4 |
Hb_001002_060 |
0.0584820156 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 5 |
Hb_025668_010 |
0.0622837162 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000803_270 |
0.0649458071 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 7 |
Hb_000006_010 |
0.0651274643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001405_210 |
0.0652107845 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 9 |
Hb_002326_040 |
0.0657729775 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 10 |
Hb_007007_040 |
0.0693629218 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 11 |
Hb_001227_120 |
0.0699633545 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 12 |
Hb_001677_200 |
0.0716656954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007248_030 |
0.0731227418 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 14 |
Hb_004586_060 |
0.0734056136 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_005730_010 |
0.0743178943 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 16 |
Hb_000963_040 |
0.0745541323 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 17 |
Hb_012506_030 |
0.0753329096 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 18 |
Hb_002014_010 |
0.0759034202 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_007413_040 |
0.0759679757 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 20 |
Hb_000085_170 |
0.0761608923 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |