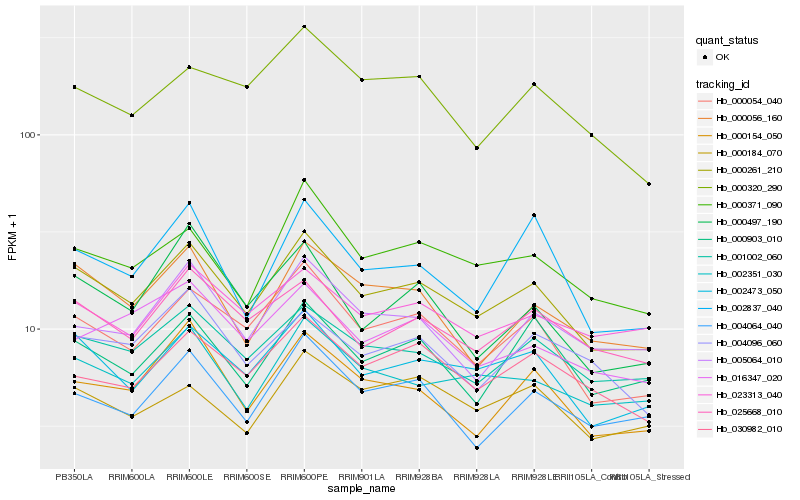
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_002473_050 |
0.0527707774 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_000054_040 |
0.0604203804 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 4 |
Hb_001002_060 |
0.0663746619 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 5 |
Hb_025668_010 |
0.0760334188 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_002351_030 |
0.0774945071 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_000371_090 |
0.0784089616 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 8 |
Hb_000056_160 |
0.0794606769 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 9 |
Hb_030982_010 |
0.0842077304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000903_010 |
0.0858824488 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 11 |
Hb_000154_050 |
0.0861645673 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 12 |
Hb_004064_040 |
0.0880827092 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 13 |
Hb_023313_040 |
0.0884564908 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000184_070 |
0.0901350835 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004096_060 |
0.0910256192 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 16 |
Hb_005064_010 |
0.0912200059 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 17 |
Hb_016347_020 |
0.0914162605 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000320_290 |
0.0916828306 |
- |
- |
PREDICTED: putative lactoylglutathione lyase [Jatropha curcas] |
| 19 |
Hb_000497_190 |
0.0924929964 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 20 |
Hb_002837_040 |
0.0935390996 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |