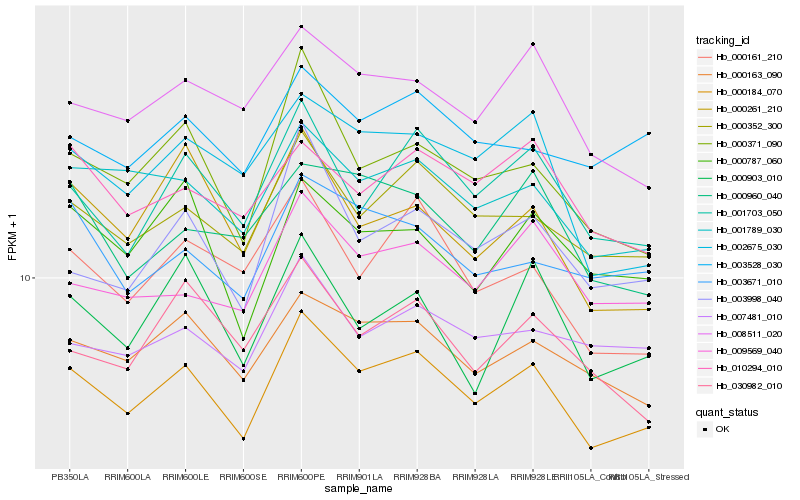
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000184_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000352_300 |
0.0648448624 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 3 |
Hb_001789_030 |
0.0718142354 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 4 |
Hb_001703_050 |
0.0727334491 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 5 |
Hb_000163_090 |
0.0728992866 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 6 |
Hb_000371_090 |
0.073671101 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 7 |
Hb_002675_030 |
0.0821372146 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 8 |
Hb_008511_020 |
0.0836973128 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 9 |
Hb_003998_040 |
0.0839293835 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 10 |
Hb_009569_040 |
0.085483612 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 11 |
Hb_000161_210 |
0.0855917592 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 12 |
Hb_003671_010 |
0.0879675325 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000787_060 |
0.088301545 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_007481_010 |
0.0894759272 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 15 |
Hb_000261_210 |
0.0901350835 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 16 |
Hb_010294_010 |
0.0909584077 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 17 |
Hb_003528_030 |
0.0913948822 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 18 |
Hb_000960_040 |
0.092097526 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 19 |
Hb_000903_010 |
0.0922145021 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 20 |
Hb_030982_010 |
0.092659435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |