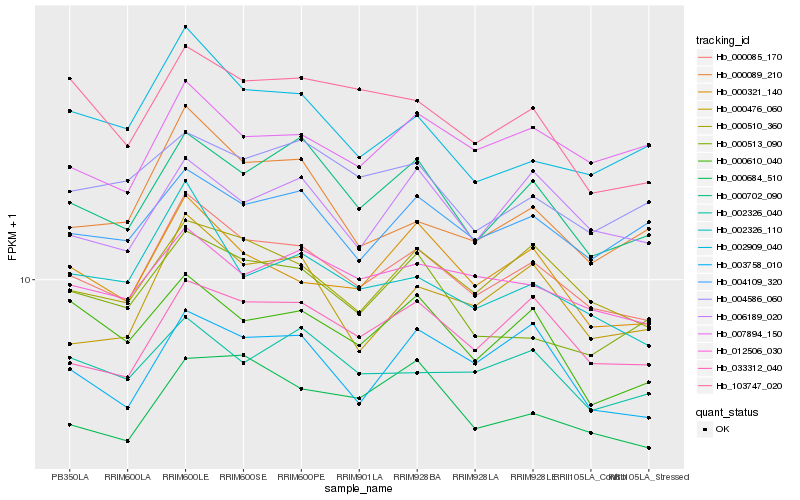
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_170 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000510_360 |
0.0504923873 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 3 |
Hb_000702_090 |
0.0573949391 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 4 |
Hb_000321_140 |
0.0595136938 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 5 |
Hb_012506_030 |
0.0604924675 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 6 |
Hb_033312_040 |
0.0608142626 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002326_110 |
0.0620214441 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 8 |
Hb_004109_320 |
0.0675402639 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 9 |
Hb_002326_040 |
0.0700721886 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 10 |
Hb_003758_010 |
0.0703226648 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_103747_020 |
0.0718978021 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 12 |
Hb_006189_020 |
0.0721015494 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 13 |
Hb_000089_210 |
0.0721019191 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_000476_060 |
0.0724008017 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000610_040 |
0.072577097 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007894_150 |
0.0727920714 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 17 |
Hb_000684_510 |
0.0729244388 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 18 |
Hb_002909_040 |
0.073617141 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 19 |
Hb_000513_090 |
0.0739231833 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 20 |
Hb_004586_060 |
0.0742456615 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |