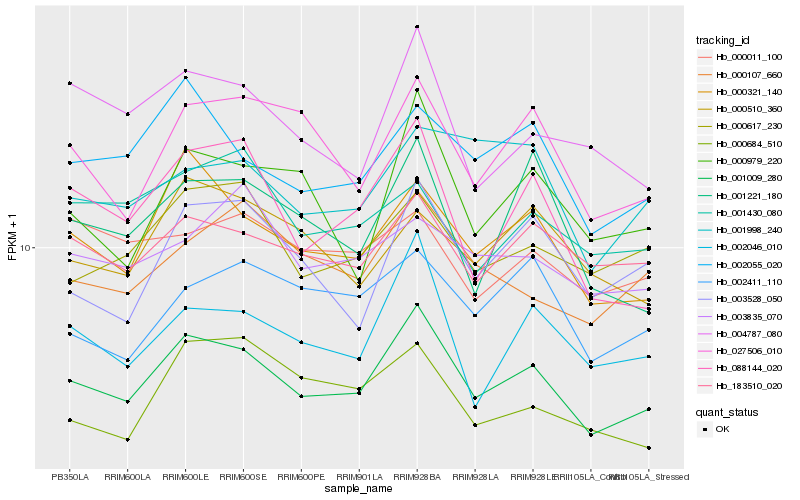
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001009_280 |
0.0 |
- |
- |
PREDICTED: telomere length regulation protein TEL2 homolog [Jatropha curcas] |
| 2 |
Hb_000321_140 |
0.0601812364 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_183510_020 |
0.0646614755 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 4 |
Hb_002055_020 |
0.0691547877 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 5 |
Hb_002046_010 |
0.0698268203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000617_230 |
0.0729031812 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 7 |
Hb_027506_010 |
0.0733855647 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 8 |
Hb_000107_660 |
0.0767320434 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 9 |
Hb_003528_050 |
0.0789965964 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_088144_020 |
0.0800476268 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002411_110 |
0.0802335198 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 12 |
Hb_004787_080 |
0.0807387579 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 13 |
Hb_000510_360 |
0.0820777958 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 14 |
Hb_000979_220 |
0.0825569382 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003835_070 |
0.0829052357 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000684_510 |
0.0837014332 |
- |
- |
run and tbc1 domain containing 3, plant, putative [Ricinus communis] |
| 17 |
Hb_000011_100 |
0.0837894481 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 18 |
Hb_001221_180 |
0.0838640411 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_001430_080 |
0.0842125832 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 20 |
Hb_001998_240 |
0.0848259361 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein-associated protein B' [Jatropha curcas] |