| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
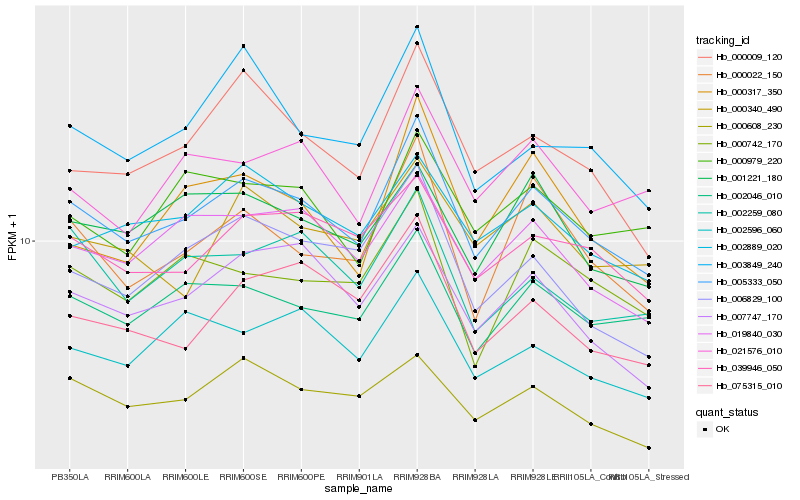
Hb_005333_050 |
0.0 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002046_010 |
0.0774268377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_019840_030 |
0.080538332 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000022_150 |
0.0830126566 |
- |
- |
- |
| 5 |
Hb_039946_050 |
0.0843171153 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_000009_120 |
0.0875794567 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 7 |
Hb_002889_020 |
0.0908387394 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 8 |
Hb_007747_170 |
0.0912427316 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 9 |
Hb_021576_010 |
0.0914033939 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 10 |
Hb_006829_100 |
0.0919922487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000742_170 |
0.0922235315 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000979_220 |
0.093198355 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002596_060 |
0.0938830993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001221_180 |
0.0941756977 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 15 |
Hb_002259_080 |
0.0945737567 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 16 |
Hb_000608_230 |
0.094775751 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 17 |
Hb_075315_010 |
0.0947843474 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 18 |
Hb_000317_350 |
0.095898317 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 19 |
Hb_000340_490 |
0.0974011955 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 20 |
Hb_003849_240 |
0.0974560319 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |