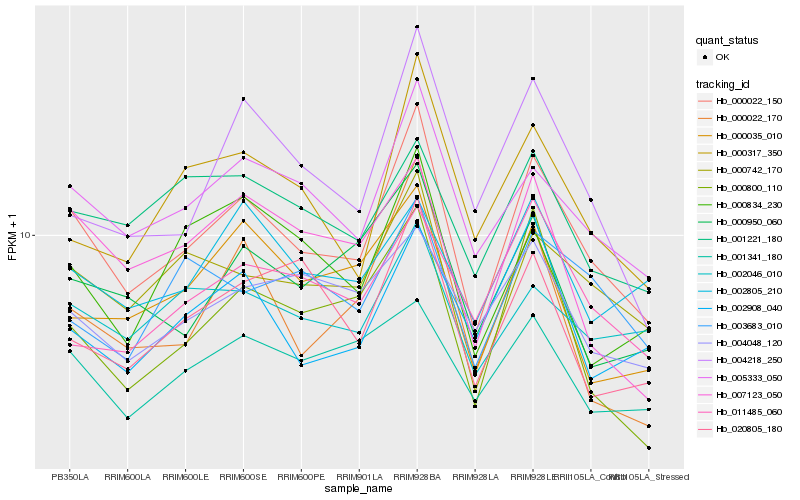
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000022_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_005333_050 |
0.0830126566 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000742_170 |
0.0908734344 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000317_350 |
0.1076386281 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 5 |
Hb_000800_110 |
0.1117175618 |
- |
- |
NAD dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_004218_250 |
0.1122553657 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 7 |
Hb_000950_060 |
0.1129400394 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 8 |
Hb_000022_170 |
0.1135441292 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 9 |
Hb_002046_010 |
0.1141467366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007123_050 |
0.1144110499 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_020805_180 |
0.1170574188 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 12 |
Hb_001341_180 |
0.1183518474 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 13 |
Hb_000035_010 |
0.1184048103 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002908_040 |
0.119035435 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_001221_180 |
0.1201183981 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_003683_010 |
0.1207860701 |
- |
- |
longevity assurance factor, putative [Ricinus communis] |
| 17 |
Hb_004048_120 |
0.1212215553 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000834_230 |
0.1213297688 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 19 |
Hb_011485_060 |
0.1231438421 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 20 |
Hb_002805_210 |
0.1247106254 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |