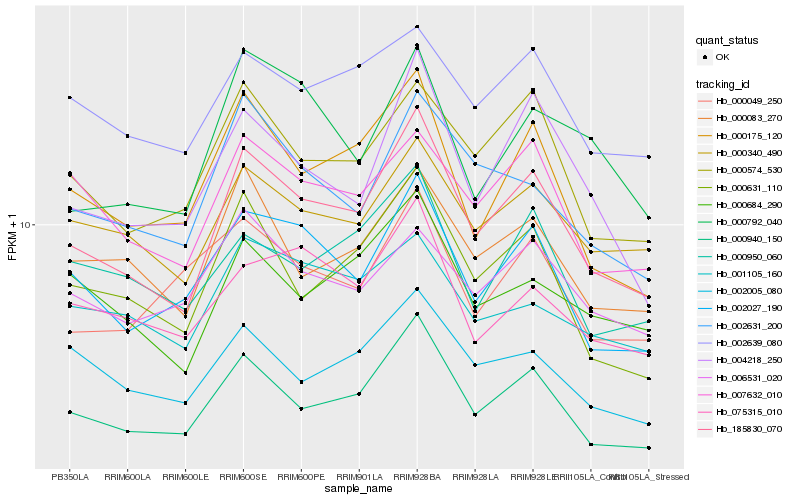
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185830_070 |
0.0 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000940_150 |
0.0650569592 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 3 |
Hb_000175_120 |
0.0900274788 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 4 |
Hb_002027_190 |
0.0900399743 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 5 |
Hb_000631_110 |
0.0918353539 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_000574_530 |
0.0921124489 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 7 |
Hb_004218_250 |
0.093555734 |
- |
- |
PREDICTED: beta-galactosidase [Jatropha curcas] |
| 8 |
Hb_000083_270 |
0.0986199166 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000340_490 |
0.0992985222 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 10 |
Hb_000684_290 |
0.1011274659 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 11 |
Hb_002631_200 |
0.1042271041 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007632_010 |
0.10475513 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002005_080 |
0.1075690841 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002639_080 |
0.1082527238 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_075315_010 |
0.1107137888 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 16 |
Hb_000950_060 |
0.1123853604 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 17 |
Hb_000792_040 |
0.1133042323 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 18 |
Hb_001105_160 |
0.1177812268 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 19 |
Hb_006531_020 |
0.1189985069 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 20 |
Hb_000049_250 |
0.119273136 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |