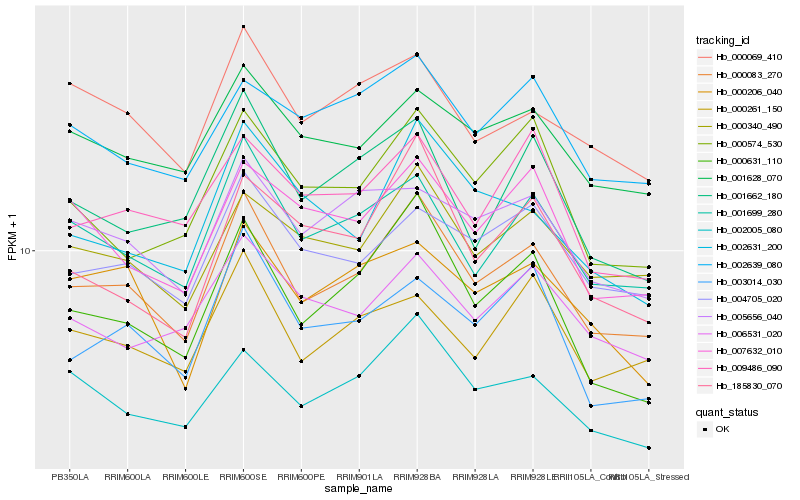
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_270 |
0.0 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001699_280 |
0.0760522929 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 3 |
Hb_000631_110 |
0.0797507599 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 4 |
Hb_000340_490 |
0.0832133704 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 5 |
Hb_002005_080 |
0.087914684 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004705_020 |
0.0887663264 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_001628_070 |
0.0916501364 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000574_530 |
0.0943439444 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 9 |
Hb_000069_410 |
0.0954248781 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 10 |
Hb_001662_180 |
0.0955933457 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 11 |
Hb_002631_200 |
0.0957310446 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005656_040 |
0.0959737395 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 13 |
Hb_000261_150 |
0.0961891385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006531_020 |
0.0963320042 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 15 |
Hb_003014_030 |
0.0977972232 |
- |
- |
PREDICTED: thyroid adenoma-associated protein homolog [Jatropha curcas] |
| 16 |
Hb_185830_070 |
0.0986199166 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_009486_090 |
0.0987502434 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_007632_010 |
0.0988795077 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002639_080 |
0.1004807059 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000206_040 |
0.101869837 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |