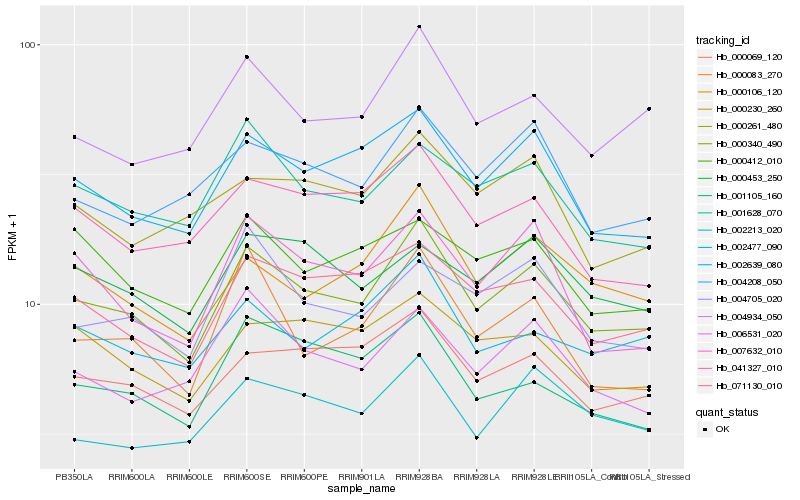
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_490 |
0.0 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 2 |
Hb_002639_080 |
0.0661587112 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_071130_010 |
0.0702198056 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004934_050 |
0.0712499389 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000069_120 |
0.07246564 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 6 |
Hb_000106_120 |
0.0747098877 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 7 |
Hb_001628_070 |
0.0766616575 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_004208_050 |
0.0795592108 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 9 |
Hb_002213_020 |
0.0810439532 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002477_090 |
0.0817784368 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007632_010 |
0.0821902125 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_041327_010 |
0.0824615292 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 13 |
Hb_006531_020 |
0.082601497 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 14 |
Hb_000083_270 |
0.0832133704 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000230_260 |
0.0835271897 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 16 |
Hb_004705_020 |
0.0851811377 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 17 |
Hb_000453_250 |
0.0860147364 |
- |
- |
PREDICTED: uncharacterized protein LOC105637185 [Jatropha curcas] |
| 18 |
Hb_001105_160 |
0.0862952134 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 19 |
Hb_000412_010 |
0.0877483613 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000261_480 |
0.0884481528 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |