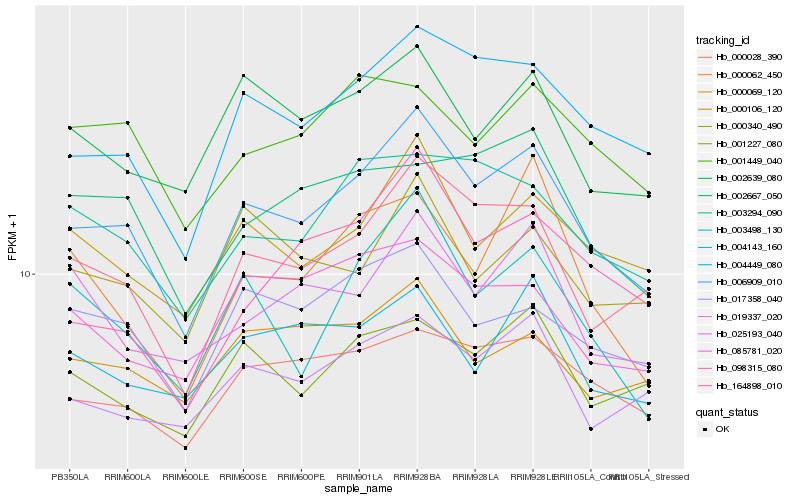
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006909_010 |
0.0 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 2 |
Hb_164898_010 |
0.0704164119 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 3 |
Hb_004449_080 |
0.0731323787 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 4 |
Hb_000106_120 |
0.0863002399 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 5 |
Hb_003498_130 |
0.0910907972 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 6 |
Hb_000028_390 |
0.0925771875 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 7 |
Hb_003294_090 |
0.0939878613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_019337_020 |
0.0948097281 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 9 |
Hb_017358_040 |
0.0951753232 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 10 |
Hb_002639_080 |
0.0955610438 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001227_080 |
0.1006247368 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002667_050 |
0.103599981 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 13 |
Hb_025193_040 |
0.1039468773 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 14 |
Hb_085781_020 |
0.104037143 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_004143_160 |
0.1041910109 |
- |
- |
SAB, putative [Ricinus communis] |
| 16 |
Hb_098315_080 |
0.1061143101 |
- |
- |
methylmalonate-semialdehyde dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000069_120 |
0.1071504097 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 18 |
Hb_000340_490 |
0.1079314697 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 19 |
Hb_001449_040 |
0.1080170733 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000062_450 |
0.1088894623 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |