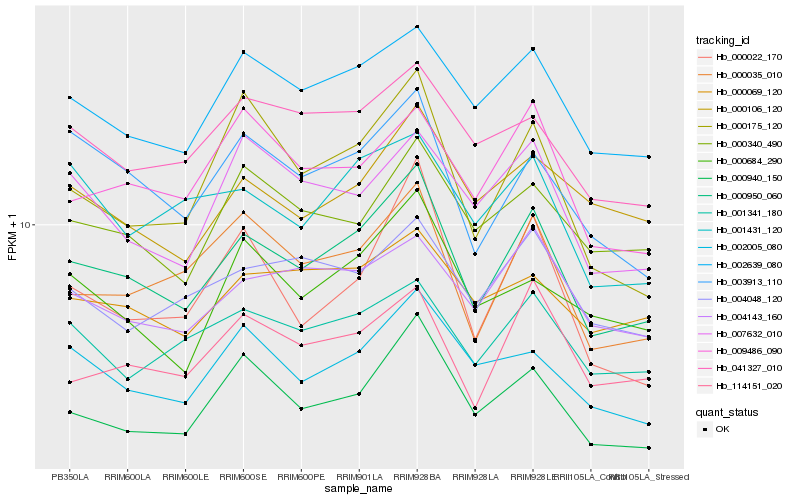
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000950_060 |
0.0 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 2 |
Hb_002639_080 |
0.0856367186 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000035_010 |
0.0859312261 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003913_110 |
0.0939740157 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 5 |
Hb_114151_020 |
0.0972752851 |
- |
- |
glucan water dikinase 2 [Manihot esculenta] |
| 6 |
Hb_000175_120 |
0.0981569427 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 7 |
Hb_001341_180 |
0.0989401239 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 8 |
Hb_002005_080 |
0.1003353358 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000940_150 |
0.1019276915 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 10 |
Hb_004143_160 |
0.1021807259 |
- |
- |
SAB, putative [Ricinus communis] |
| 11 |
Hb_009486_090 |
0.103017635 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_041327_010 |
0.1031232891 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 13 |
Hb_004048_120 |
0.1036028568 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000340_490 |
0.1047758508 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 15 |
Hb_000684_290 |
0.1056353589 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 16 |
Hb_001431_120 |
0.1069590712 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 17 |
Hb_000069_120 |
0.1076391946 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 18 |
Hb_000106_120 |
0.1083299064 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 19 |
Hb_000022_170 |
0.1094023488 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 20 |
Hb_007632_010 |
0.1094859475 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |