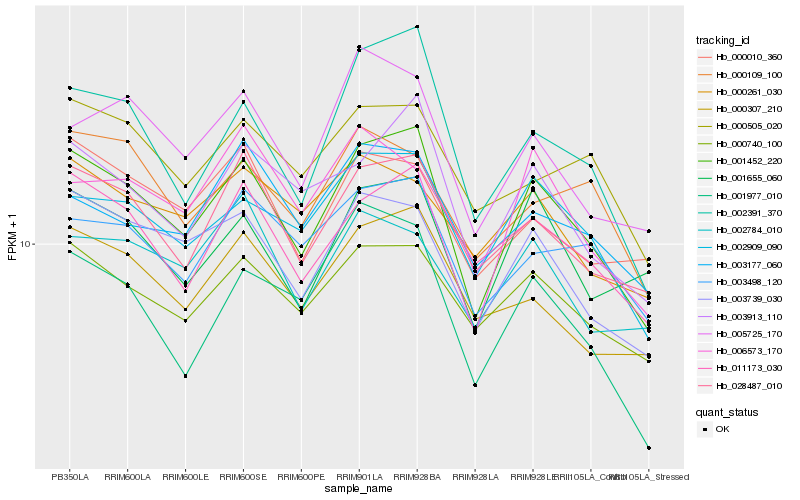
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001452_220 |
0.0 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 2 |
Hb_003739_030 |
0.0733446084 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 3 |
Hb_002391_370 |
0.0773546096 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 4 |
Hb_003913_110 |
0.0791344715 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 5 |
Hb_028487_010 |
0.08040431 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002909_090 |
0.0826047546 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 7 |
Hb_011173_030 |
0.0862966 |
- |
- |
pumilio, putative [Ricinus communis] |
| 8 |
Hb_000740_100 |
0.086381496 |
- |
- |
calpain, putative [Ricinus communis] |
| 9 |
Hb_005725_170 |
0.0926313412 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X2 [Jatropha curcas] |
| 10 |
Hb_000307_210 |
0.0932227798 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_001655_060 |
0.0933731302 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 12 |
Hb_003498_120 |
0.0949703751 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 13 |
Hb_001977_010 |
0.0999296421 |
- |
- |
PREDICTED: uncharacterized protein LOC105636485 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000505_020 |
0.101357065 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 15 |
Hb_000010_360 |
0.1015427408 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 16 |
Hb_006573_170 |
0.1024168982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003177_060 |
0.1046211865 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002784_010 |
0.1046451795 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 19 |
Hb_000109_100 |
0.1046473105 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 20 |
Hb_000261_030 |
0.1060506208 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |