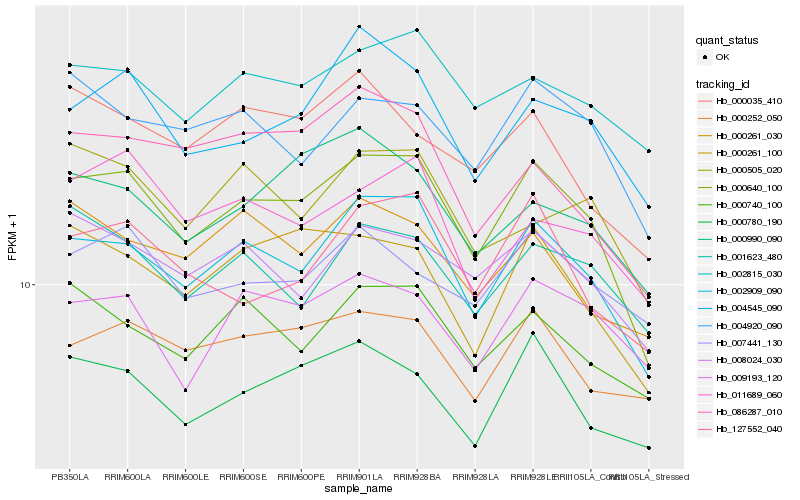
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_100 |
0.0 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_002909_090 |
0.0494317141 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 3 |
Hb_000261_100 |
0.0840508791 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004545_090 |
0.0912073132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000780_190 |
0.0918534635 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 6 |
Hb_008024_030 |
0.0925932033 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 7 |
Hb_127552_040 |
0.0929990265 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 8 |
Hb_002815_030 |
0.0935697783 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 9 |
Hb_004920_090 |
0.0937655982 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000740_100 |
0.0947199464 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_000252_050 |
0.0972507344 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 12 |
Hb_009193_120 |
0.0985087702 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 13 |
Hb_000990_090 |
0.0992885503 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_001623_480 |
0.1005336717 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 13 [Jatropha curcas] |
| 15 |
Hb_000035_410 |
0.1021579634 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 16 |
Hb_011689_060 |
0.1026389965 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 17 |
Hb_086287_010 |
0.1027268987 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007441_130 |
0.1033302035 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000261_030 |
0.103535361 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 20 |
Hb_000505_020 |
0.1036816086 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |