| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
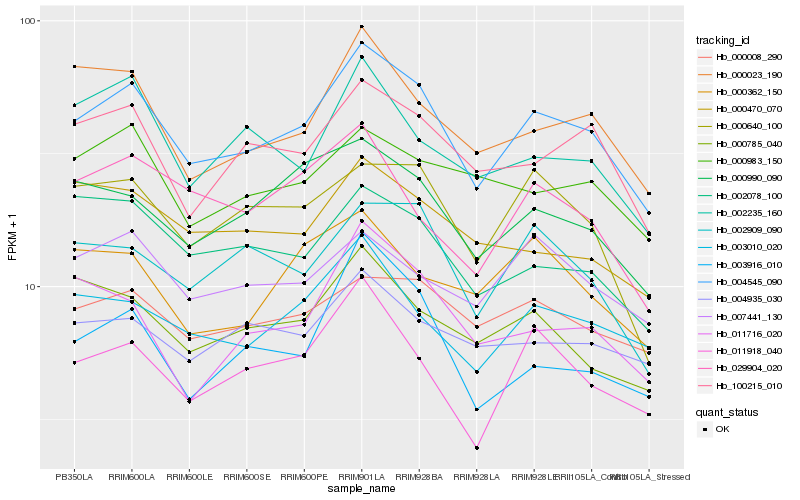
Hb_004545_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_100215_010 |
0.0810881359 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 3 |
Hb_003010_020 |
0.0818699176 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000990_090 |
0.0836310298 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_000023_190 |
0.0839679972 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 6 |
Hb_011918_040 |
0.0860629455 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 7 |
Hb_002909_090 |
0.0877983597 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase DPE2 [Jatropha curcas] |
| 8 |
Hb_000008_290 |
0.0896191498 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 9 |
Hb_000640_100 |
0.0912073132 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002078_100 |
0.0921266255 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 11 |
Hb_000785_040 |
0.0922650084 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 12 |
Hb_007441_130 |
0.0943624061 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_004935_030 |
0.0947283577 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 14 |
Hb_000983_150 |
0.09619833 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 15 |
Hb_011716_020 |
0.0965268071 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 16 |
Hb_000470_070 |
0.0971418788 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 17 |
Hb_002235_160 |
0.0981850409 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 18 |
Hb_029904_020 |
0.0984931905 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 19 |
Hb_003916_010 |
0.0998421819 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 20 |
Hb_000362_150 |
0.1001277228 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |