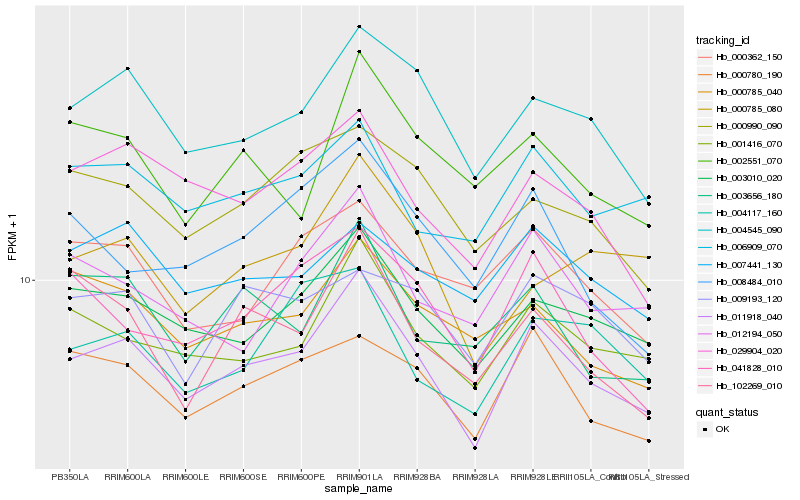
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011918_040 |
0.0 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 2 |
Hb_003010_020 |
0.0838173049 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_001416_070 |
0.0844520978 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_004545_090 |
0.0860629455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000780_190 |
0.0972297891 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 6 |
Hb_029904_020 |
0.1017634255 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 7 |
Hb_041828_010 |
0.106045694 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 8 |
Hb_006909_070 |
0.1075429019 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_012194_050 |
0.1079619204 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 10 |
Hb_000990_090 |
0.1090623241 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_000785_040 |
0.1110842937 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 12 |
Hb_003656_180 |
0.1117640094 |
- |
- |
PREDICTED: uncharacterized protein LOC105637927 [Jatropha curcas] |
| 13 |
Hb_002551_070 |
0.1136337387 |
- |
- |
hypothetical protein POPTR_0019s10130g [Populus trichocarpa] |
| 14 |
Hb_000362_150 |
0.1138637647 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 15 |
Hb_000785_080 |
0.1139508483 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 16 |
Hb_007441_130 |
0.1150485946 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_009193_120 |
0.1152297832 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 18 |
Hb_008484_010 |
0.1152309395 |
- |
- |
hypothetical protein B456_008G050700 [Gossypium raimondii] |
| 19 |
Hb_004117_160 |
0.116429629 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |
| 20 |
Hb_102269_010 |
0.1165555037 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |