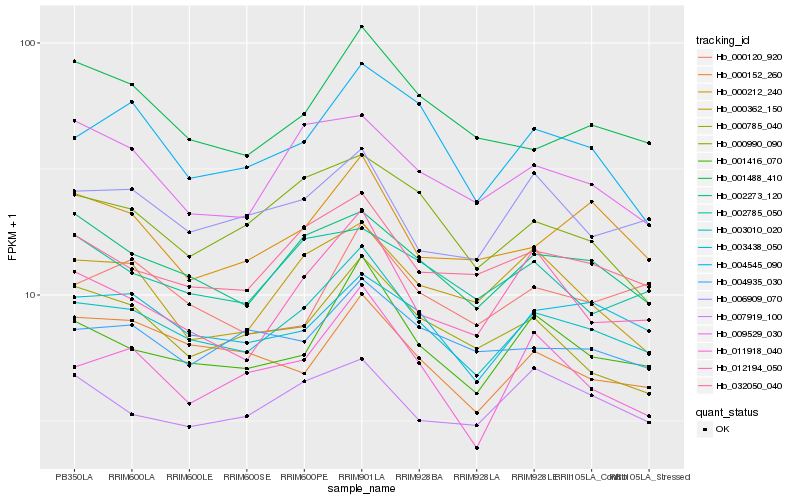
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003010_020 |
0.0 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001416_070 |
0.0587604388 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 3 |
Hb_032050_040 |
0.0659797841 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 4 |
Hb_002273_120 |
0.067234594 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 5 |
Hb_003438_050 |
0.0771258072 |
- |
- |
PREDICTED: rab3 GTPase-activating protein catalytic subunit isoform X2 [Jatropha curcas] |
| 6 |
Hb_012194_050 |
0.0812469515 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 7 |
Hb_006909_070 |
0.0815971654 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_004545_090 |
0.0818699176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001488_410 |
0.082297651 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 10 |
Hb_000212_240 |
0.083545695 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_011918_040 |
0.0838173049 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 12 |
Hb_004935_030 |
0.0841523077 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 13 |
Hb_009529_030 |
0.0843078267 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 14 |
Hb_000990_090 |
0.085295206 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 15 |
Hb_002785_050 |
0.0872733907 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 16 |
Hb_000785_040 |
0.0881560371 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 17 |
Hb_000120_920 |
0.0887861512 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 18 |
Hb_000152_260 |
0.0891150875 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 19 |
Hb_000362_150 |
0.0894172167 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 20 |
Hb_007919_100 |
0.0897570636 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |