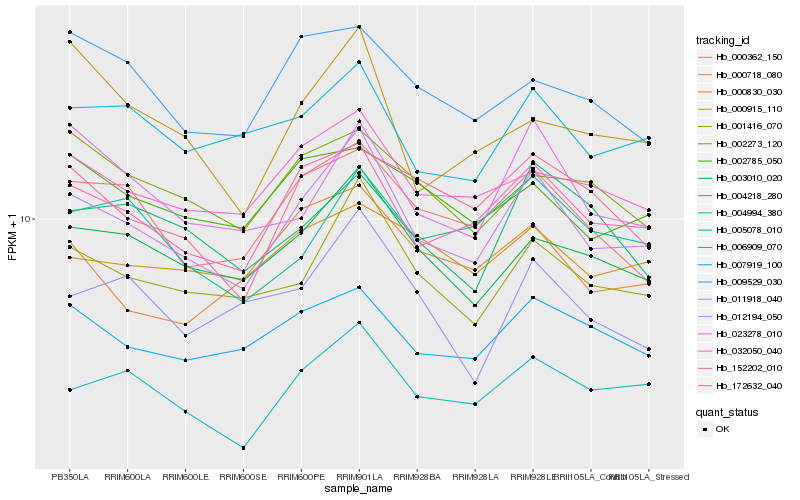
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012194_050 |
0.0 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 2 |
Hb_152202_010 |
0.0672305593 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 3 |
Hb_001416_070 |
0.0706813135 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 4 |
Hb_005078_010 |
0.0798691409 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 5 |
Hb_003010_020 |
0.0812469515 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000362_150 |
0.0861125559 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 7 |
Hb_032050_040 |
0.0885432829 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 8 |
Hb_007919_100 |
0.0928366663 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 9 |
Hb_006909_070 |
0.0955996301 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_023278_010 |
0.0992775793 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 11 |
Hb_004994_380 |
0.1060917346 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 12 |
Hb_172632_040 |
0.1066947574 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 13 |
Hb_000718_080 |
0.1072679057 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |
| 14 |
Hb_011918_040 |
0.1079619204 |
- |
- |
PREDICTED: dnaJ protein homolog 2 [Jatropha curcas] |
| 15 |
Hb_000830_030 |
0.1081430175 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 16 |
Hb_002273_120 |
0.1083049341 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 17 |
Hb_002785_050 |
0.1098258 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 18 |
Hb_009529_030 |
0.1112225737 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 19 |
Hb_004218_280 |
0.1120627637 |
- |
- |
Exoenzymes regulatory protein aepA precursor, putative [Ricinus communis] |
| 20 |
Hb_000915_110 |
0.114565481 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |