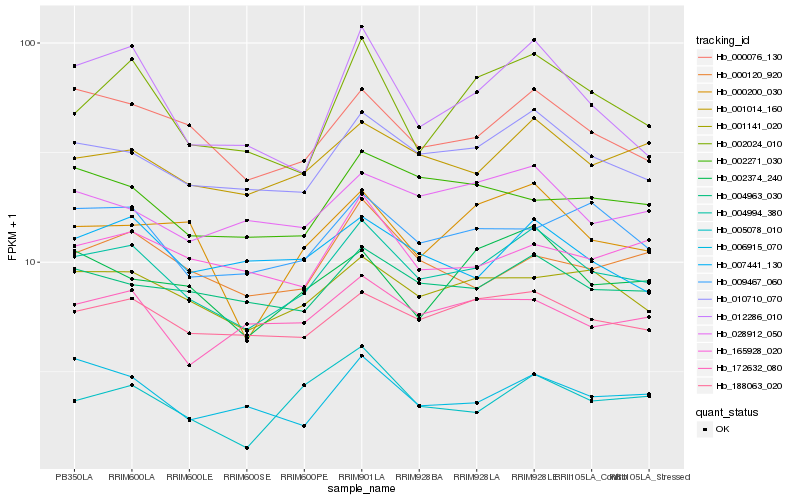
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_380 |
0.0 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 2 |
Hb_010710_070 |
0.0576055766 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_000076_130 |
0.0733872931 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 4 |
Hb_001014_160 |
0.0760779794 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 5 |
Hb_188063_020 |
0.0777716863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001141_020 |
0.07918324 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 7 |
Hb_004963_030 |
0.0801542252 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_009467_060 |
0.089929873 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000200_030 |
0.0918954844 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002024_010 |
0.0924538529 |
- |
- |
hypothetical protein B456_003G0564002, partial [Gossypium raimondii] |
| 11 |
Hb_028912_050 |
0.0931236869 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 12 |
Hb_172632_080 |
0.0936410718 |
- |
- |
PREDICTED: uncharacterized protein LOC105141138 isoform X1 [Populus euphratica] |
| 13 |
Hb_002374_240 |
0.0937682342 |
- |
- |
nucellin, putative [Ricinus communis] |
| 14 |
Hb_000120_920 |
0.0946376332 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 15 |
Hb_006915_070 |
0.0955589315 |
- |
- |
hypothetical protein CAPTEDRAFT_111329, partial [Capitella teleta] |
| 16 |
Hb_007441_130 |
0.0957112025 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_005078_010 |
0.0975074722 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 18 |
Hb_165928_020 |
0.0975705988 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_012286_010 |
0.0981172647 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit C-like [Jatropha curcas] |
| 20 |
Hb_002271_030 |
0.0992211801 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |