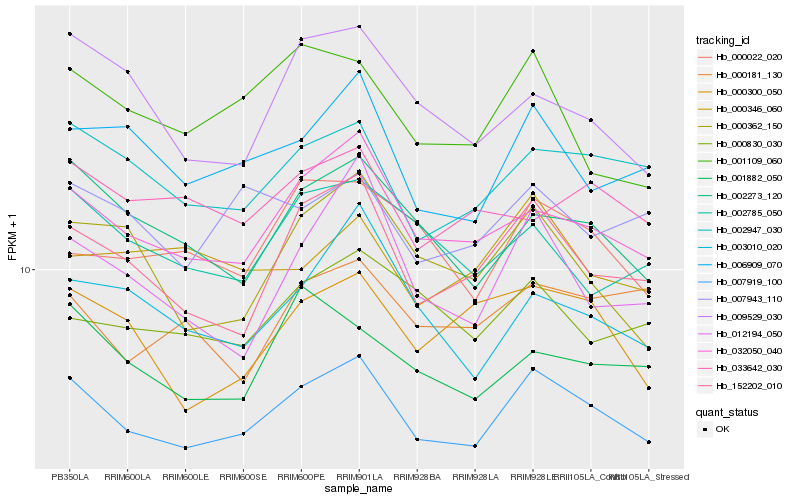
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007919_100 |
0.0 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 2 |
Hb_032050_040 |
0.0607994599 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 3 |
Hb_152202_010 |
0.066949718 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 4 |
Hb_002947_030 |
0.0702769275 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 5 |
Hb_006909_070 |
0.0777332241 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_002273_120 |
0.0787388695 |
- |
- |
PREDICTED: probable apyrase 6 [Jatropha curcas] |
| 7 |
Hb_000300_050 |
0.0862183124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000022_020 |
0.0870149853 |
- |
- |
hypothetical protein CICLE_v10020565mg [Citrus clementina] |
| 9 |
Hb_007943_110 |
0.0873929517 |
- |
- |
PREDICTED: basic salivary proline-rich protein 3 [Jatropha curcas] |
| 10 |
Hb_000830_030 |
0.0878231575 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 11 |
Hb_001109_060 |
0.0887801104 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_002785_050 |
0.0892006176 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 13 |
Hb_003010_020 |
0.0897570636 |
- |
- |
PREDICTED: probable inactive heme oxygenase 2, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001882_050 |
0.0904557002 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000362_150 |
0.0917331971 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 16 |
Hb_000181_130 |
0.0919162171 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_012194_050 |
0.0928366663 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 18 |
Hb_009529_030 |
0.0931609594 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 19 |
Hb_033642_030 |
0.0935254547 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000346_060 |
0.0945496778 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |