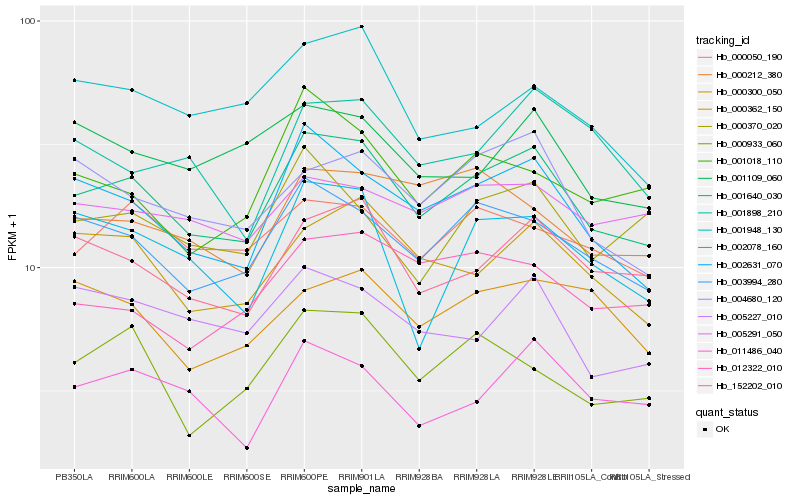
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001640_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 2 |
Hb_002631_070 |
0.0717664555 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 3 |
Hb_003994_280 |
0.0737511417 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 4 |
Hb_152202_010 |
0.0814769034 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 5 |
Hb_000362_150 |
0.086700503 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 6 |
Hb_004680_120 |
0.0953130345 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 7 |
Hb_000050_190 |
0.096462639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011486_040 |
0.0998669757 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 9 |
Hb_005227_010 |
0.1010964301 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005291_050 |
0.1020332543 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002078_160 |
0.1032863281 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 12 |
Hb_001018_110 |
0.103651415 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_000370_020 |
0.1045650777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001109_060 |
0.104871091 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000933_060 |
0.1054851109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012322_010 |
0.1055224715 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000212_380 |
0.1056947907 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 18 |
Hb_001948_130 |
0.1063414024 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 19 |
Hb_001898_210 |
0.1066413734 |
- |
- |
PREDICTED: shaggy-related protein kinase kappa [Populus euphratica] |
| 20 |
Hb_000300_050 |
0.1068162388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |