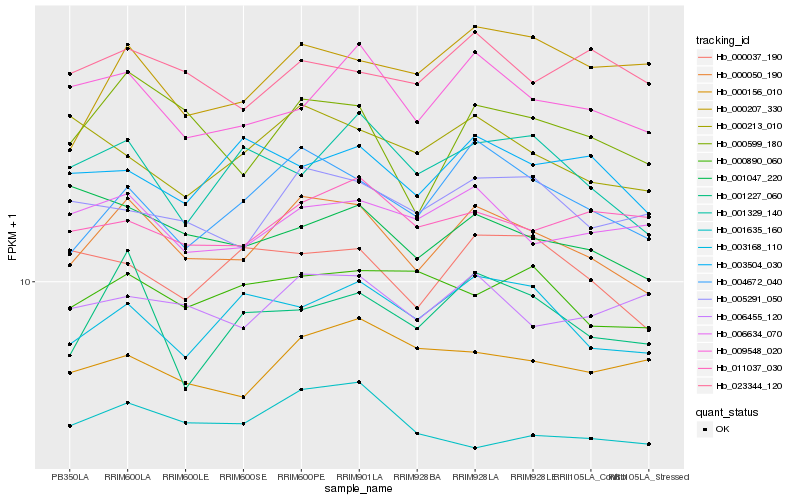
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000050_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004672_040 |
0.0670886875 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 3 |
Hb_000599_180 |
0.0709040197 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_009548_020 |
0.0798517172 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 5 |
Hb_005291_050 |
0.0823535188 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003168_110 |
0.0824048021 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_006634_070 |
0.0833073834 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 8 |
Hb_006455_120 |
0.0849478864 |
- |
- |
n6-DNA-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001227_060 |
0.0865492907 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20740 [Jatropha curcas] |
| 10 |
Hb_000037_190 |
0.0872902998 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 11 |
Hb_001635_160 |
0.0886603425 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 12 |
Hb_000890_060 |
0.0888047781 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 13 |
Hb_000156_010 |
0.0898577655 |
- |
- |
phospho-n-acetylmuramoyl-pentapeptide-transferase, putative [Ricinus communis] |
| 14 |
Hb_023344_120 |
0.0902638711 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001329_140 |
0.0916672769 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 16 |
Hb_011037_030 |
0.0918738627 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 17 |
Hb_000213_010 |
0.0921322674 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 18 |
Hb_001047_220 |
0.093094558 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 19 |
Hb_003504_030 |
0.0939976657 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000207_330 |
0.0944667602 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |