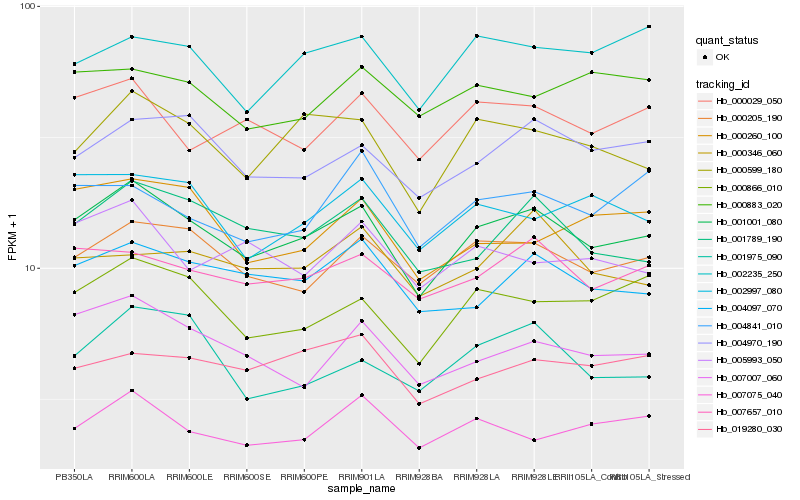
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001001_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 2 |
Hb_000599_180 |
0.0629991023 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000205_190 |
0.0639959088 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_007657_010 |
0.0674324803 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_004097_070 |
0.0686095052 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 6 |
Hb_001789_190 |
0.0695034685 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 7 |
Hb_004970_190 |
0.0698976636 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000866_010 |
0.0707648539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007007_060 |
0.0721731489 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000029_050 |
0.075386268 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 11 |
Hb_002997_080 |
0.0774485556 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 12 |
Hb_002235_250 |
0.080378054 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_019280_030 |
0.0804166328 |
- |
- |
- |
| 14 |
Hb_007075_040 |
0.0831436324 |
- |
- |
hypothetical protein CISIN_1g039518mg, partial [Citrus sinensis] |
| 15 |
Hb_005993_050 |
0.0843412862 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 16 |
Hb_000260_100 |
0.0852890981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000883_020 |
0.0861051077 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 18 |
Hb_001975_090 |
0.0861114191 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 19 |
Hb_000346_060 |
0.0872446885 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 20 |
Hb_004841_010 |
0.0887634697 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |