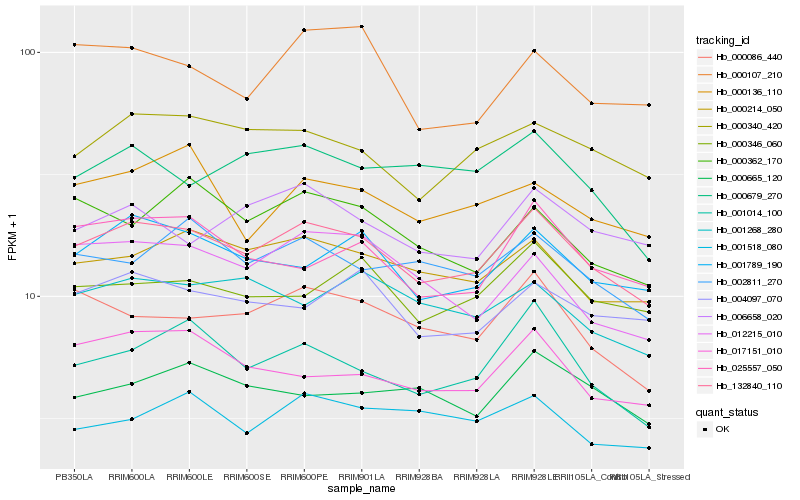
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_110 |
0.0 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000214_050 |
0.0645649057 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001789_190 |
0.067770742 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 4 |
Hb_000340_420 |
0.0689004163 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_000086_440 |
0.0718747226 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 6 |
Hb_001014_100 |
0.0749963172 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 7 |
Hb_002811_270 |
0.0753258444 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 8 |
Hb_000346_060 |
0.0760839106 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 9 |
Hb_004097_070 |
0.0770646988 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 10 |
Hb_025557_050 |
0.0776963189 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 11 |
Hb_006658_020 |
0.0786142279 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 12 |
Hb_017151_010 |
0.0789002262 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 13 |
Hb_000679_270 |
0.0820060413 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 14 |
Hb_001268_280 |
0.0823101991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000362_170 |
0.0827045434 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 16 |
Hb_000136_110 |
0.0827278968 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 17 |
Hb_012215_010 |
0.0837553925 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 18 |
Hb_000665_120 |
0.0837588198 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 19 |
Hb_001518_080 |
0.0847952049 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 20 |
Hb_000107_210 |
0.0853636713 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |