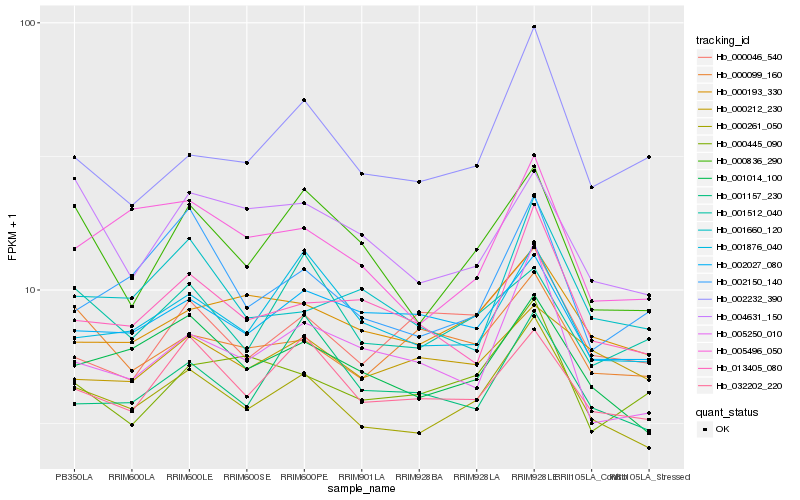
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_050 |
0.0 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 2 |
Hb_001014_100 |
0.0697395136 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 3 |
Hb_032202_220 |
0.0869953666 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_000193_330 |
0.0917432892 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 5 |
Hb_005496_050 |
0.0917771553 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_001876_040 |
0.0941165472 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 7 |
Hb_002027_080 |
0.0962569886 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 8 |
Hb_000836_290 |
0.097069507 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_013405_080 |
0.0973269004 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 10 |
Hb_000099_160 |
0.0999966621 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 11 |
Hb_001157_230 |
0.1037095339 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 12 |
Hb_005250_010 |
0.1044957676 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 13 |
Hb_002232_390 |
0.1046394581 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 14 |
Hb_001512_040 |
0.10472363 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 15 |
Hb_000445_090 |
0.1057965353 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 16 |
Hb_002150_140 |
0.1061184988 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 17 |
Hb_000046_540 |
0.106266623 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 18 |
Hb_001660_120 |
0.1065266058 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000212_230 |
0.1070709127 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 20 |
Hb_004631_150 |
0.1090092422 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |