| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
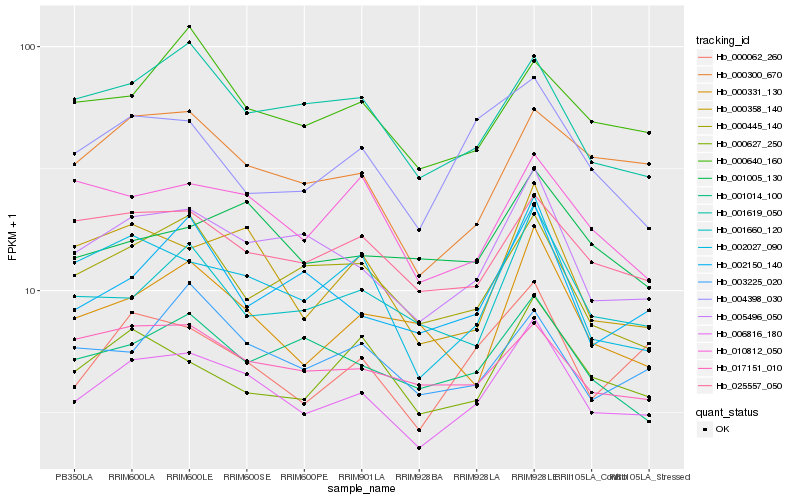
Hb_006816_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_005496_050 |
0.0717761726 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 3 |
Hb_000062_260 |
0.0897314038 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 4 |
Hb_000358_140 |
0.0969086843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002027_090 |
0.0970859458 |
- |
- |
- |
| 6 |
Hb_025557_050 |
0.0981145575 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 7 |
Hb_000300_670 |
0.1000898246 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001619_050 |
0.1025422143 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001660_120 |
0.1027697743 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001005_130 |
0.1048275869 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 11 |
Hb_001014_100 |
0.1057292913 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 12 |
Hb_000627_250 |
0.1059023384 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 13 |
Hb_017151_010 |
0.1078092214 |
- |
- |
PREDICTED: uncharacterized protein LOC105641966 [Jatropha curcas] |
| 14 |
Hb_000640_160 |
0.1089036553 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 15 |
Hb_000445_140 |
0.1089657359 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 16 |
Hb_004398_030 |
0.1097507983 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 17 |
Hb_000331_130 |
0.1102840721 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002150_140 |
0.1118463118 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 19 |
Hb_010812_050 |
0.1120632067 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003225_020 |
0.1126396423 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |