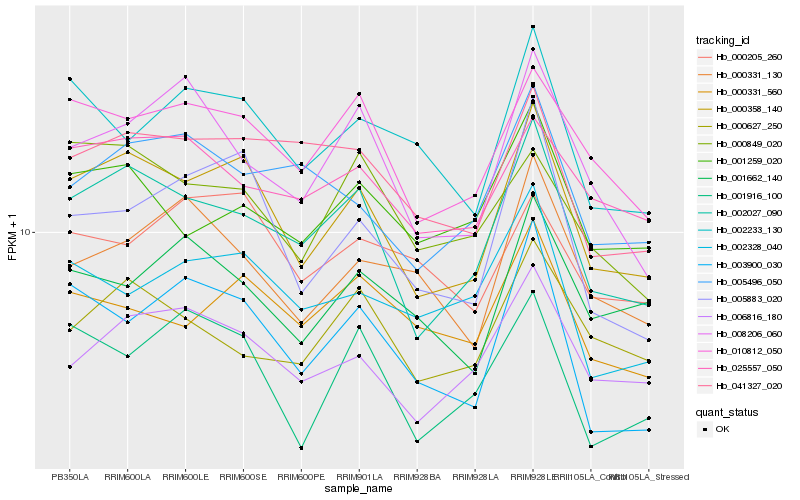
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000358_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002027_090 |
0.0699665875 |
- |
- |
- |
| 3 |
Hb_005883_020 |
0.0930846952 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 4 |
Hb_010812_050 |
0.0959554436 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006816_180 |
0.0969086843 |
- |
- |
- |
| 6 |
Hb_041327_020 |
0.1081487722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002233_130 |
0.1113421126 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_003900_030 |
0.1116589189 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000331_560 |
0.1141225312 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 10 |
Hb_001259_020 |
0.1142258811 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000849_020 |
0.1149713403 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001916_100 |
0.1157222187 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g09950 [Jatropha curcas] |
| 13 |
Hb_008206_060 |
0.1159535212 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_025557_050 |
0.116996136 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 15 |
Hb_005496_050 |
0.1177744488 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_001662_140 |
0.1186468548 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002328_040 |
0.1189445556 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000331_130 |
0.1201756893 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000205_260 |
0.1219884147 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 20 |
Hb_000627_250 |
0.122161905 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |