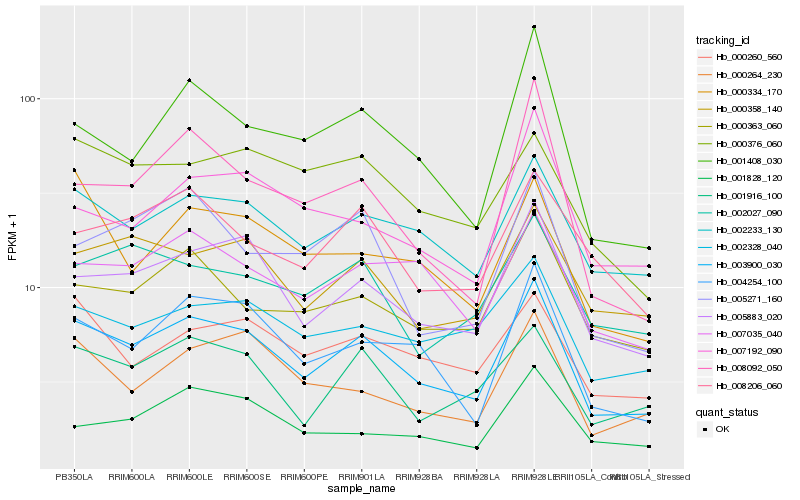
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003900_030 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005883_020 |
0.08570752 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002233_130 |
0.0907729127 |
- |
- |
transporter, putative [Ricinus communis] |
| 4 |
Hb_004254_100 |
0.0965056184 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007035_040 |
0.1093641306 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001916_100 |
0.1094481763 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g09950 [Jatropha curcas] |
| 7 |
Hb_000363_060 |
0.1106490036 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000358_140 |
0.1116589189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001408_030 |
0.1129980417 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002328_040 |
0.1149304178 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000264_230 |
0.1151084195 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 12 |
Hb_000260_560 |
0.1158191205 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008206_060 |
0.1219774985 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_002027_090 |
0.1228437095 |
- |
- |
- |
| 15 |
Hb_000376_060 |
0.1238533188 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005271_160 |
0.1242380582 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |
| 17 |
Hb_000334_170 |
0.1245485569 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007192_090 |
0.1255541616 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 19 |
Hb_008092_050 |
0.1259620958 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 20 |
Hb_001828_120 |
0.1278523459 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |