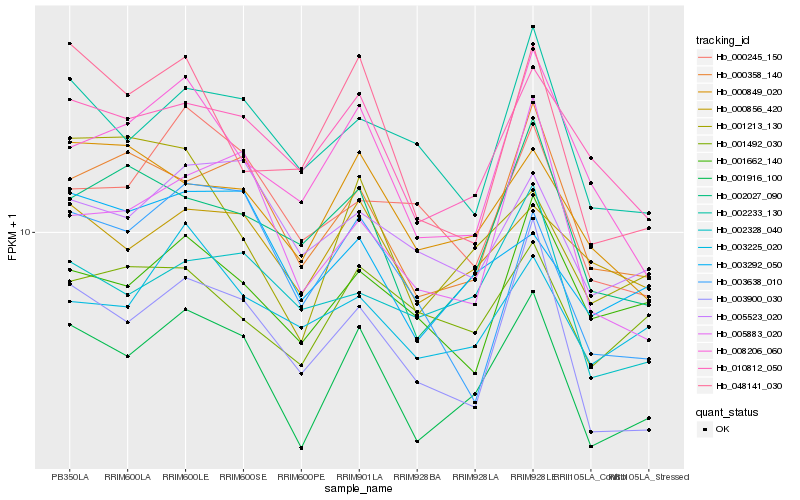
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001916_100 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g09950 [Jatropha curcas] |
| 2 |
Hb_003900_030 |
0.1094481763 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000358_140 |
0.1157222187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010812_050 |
0.126583836 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At5g05200, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002027_090 |
0.1303902175 |
- |
- |
- |
| 6 |
Hb_005883_020 |
0.1318600615 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002233_130 |
0.1321683616 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_003292_050 |
0.1325424401 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 9 |
Hb_008206_060 |
0.1326429282 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_048141_030 |
0.1341406251 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g59040-like [Prunus mume] |
| 11 |
Hb_000856_420 |
0.1352296087 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_005523_020 |
0.135599904 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 13 |
Hb_000849_020 |
0.1358604135 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002328_040 |
0.1397780231 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003638_010 |
0.1411453698 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 16 |
Hb_003225_020 |
0.1415446779 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 17 |
Hb_001213_130 |
0.1423014844 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001662_140 |
0.1423415478 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001492_030 |
0.1442363648 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000245_150 |
0.1447204348 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |