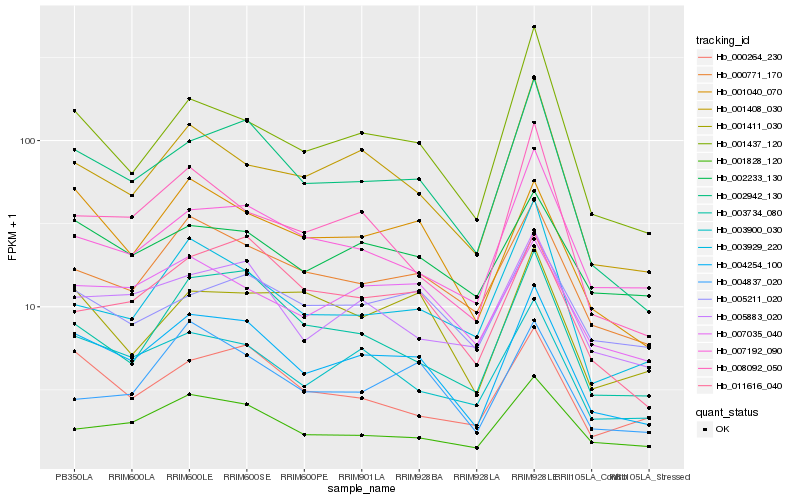
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004254_100 |
0.0 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002942_130 |
0.0782994315 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 3 |
Hb_003900_030 |
0.0965056184 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000771_170 |
0.1043076288 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_001040_070 |
0.1049097467 |
- |
- |
PREDICTED: uncharacterized protein LOC105649523 [Jatropha curcas] |
| 6 |
Hb_001408_030 |
0.1099959668 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005883_020 |
0.1103215664 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003734_080 |
0.1158563649 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 9 |
Hb_007035_040 |
0.1162308363 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011616_040 |
0.1183480926 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 11 |
Hb_001828_120 |
0.1184314011 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 12 |
Hb_001437_120 |
0.1192354535 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 13 |
Hb_000264_230 |
0.1197758148 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 14 |
Hb_007192_090 |
0.1213530845 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 15 |
Hb_002233_130 |
0.12217425 |
- |
- |
transporter, putative [Ricinus communis] |
| 16 |
Hb_008092_050 |
0.1281466689 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 17 |
Hb_004837_020 |
0.1291017109 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 18 |
Hb_001411_030 |
0.1316017188 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 19 |
Hb_003929_220 |
0.1321615872 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005211_020 |
0.1326656587 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |