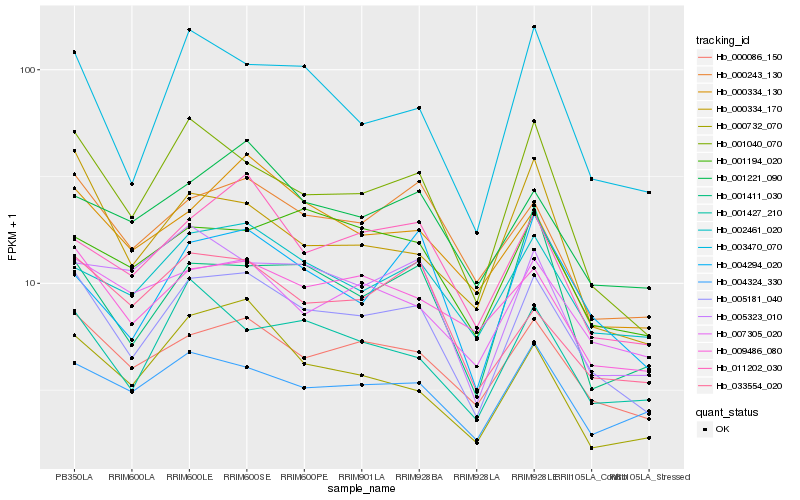
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005181_040 |
0.0 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 2 |
Hb_000086_150 |
0.0817327135 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001040_070 |
0.0886242638 |
- |
- |
PREDICTED: uncharacterized protein LOC105649523 [Jatropha curcas] |
| 4 |
Hb_000243_130 |
0.0999893962 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 5 |
Hb_003470_070 |
0.102001873 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 6 |
Hb_009486_080 |
0.1036158658 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 7 |
Hb_002461_020 |
0.1044953063 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001427_210 |
0.1094993986 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011202_030 |
0.1095494611 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 10 |
Hb_007305_020 |
0.1097165682 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 11 |
Hb_001221_090 |
0.1126198823 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000334_130 |
0.1134042668 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 13 |
Hb_004324_330 |
0.1144898354 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 14 |
Hb_004294_020 |
0.1163346067 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001194_020 |
0.1173792172 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 16 |
Hb_033554_020 |
0.1183288492 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 17 |
Hb_000732_070 |
0.118961215 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 18 |
Hb_005323_010 |
0.1195818277 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 19 |
Hb_000334_170 |
0.1206787029 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001411_030 |
0.1208353172 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |