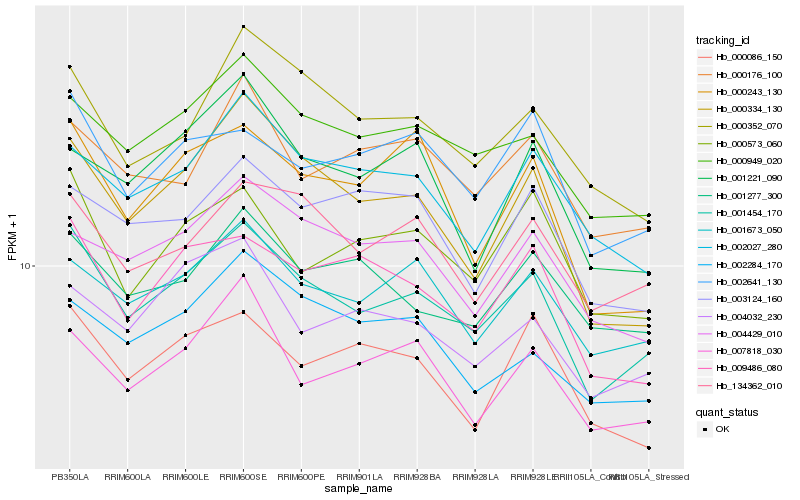
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 2 |
Hb_000949_020 |
0.0697892975 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 3 |
Hb_001673_050 |
0.0770726674 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000334_130 |
0.0805986736 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 5 |
Hb_134362_010 |
0.0806235627 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 6 |
Hb_009486_080 |
0.082302081 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 7 |
Hb_000573_060 |
0.0864070827 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 8 |
Hb_002641_130 |
0.0868909764 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 9 |
Hb_000352_070 |
0.0873839416 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 10 |
Hb_002027_280 |
0.0887369489 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 11 |
Hb_001277_300 |
0.0903492499 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 12 |
Hb_007818_030 |
0.0917526827 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 13 |
Hb_000243_130 |
0.0918543393 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 14 |
Hb_004032_230 |
0.0924091011 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |
| 15 |
Hb_004429_010 |
0.0927844382 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_000176_100 |
0.0936448754 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 17 |
Hb_002284_170 |
0.0946365016 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 18 |
Hb_000086_150 |
0.0953630165 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003124_160 |
0.0981938218 |
- |
- |
dynamin, putative [Ricinus communis] |
| 20 |
Hb_001221_090 |
0.1003508161 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |