| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
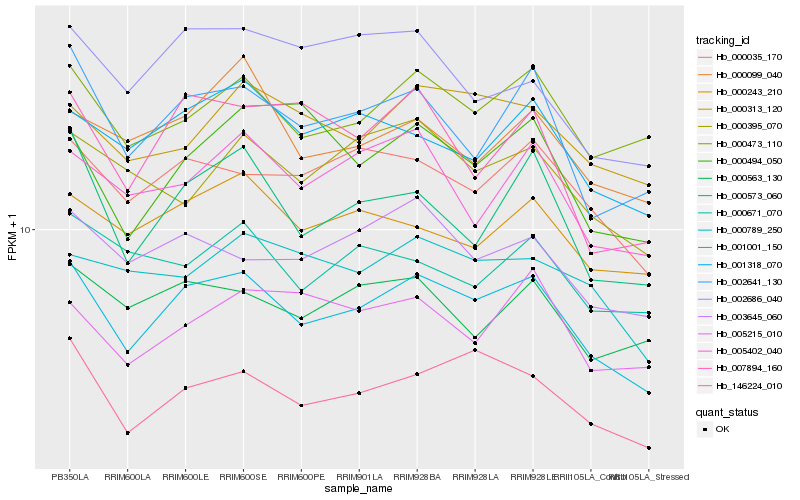
Hb_001001_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_146224_010 |
0.0695759291 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002641_130 |
0.0727272254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 4 |
Hb_000035_170 |
0.0755112772 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 5 |
Hb_000573_060 |
0.0782372009 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 6 |
Hb_000473_110 |
0.0782382633 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 7 |
Hb_001318_070 |
0.0823489448 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003645_060 |
0.0837302 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005402_040 |
0.0867419046 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 10 |
Hb_000243_210 |
0.0873663782 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000313_120 |
0.0880459081 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 12 |
Hb_000563_130 |
0.0906582955 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 13 |
Hb_007894_160 |
0.0908497944 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 14 |
Hb_000395_070 |
0.0915710441 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000099_040 |
0.0917777455 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_000789_250 |
0.0929371803 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 17 |
Hb_002686_040 |
0.0937236294 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 18 |
Hb_005215_010 |
0.0949458448 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000494_050 |
0.0971246705 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_000671_070 |
0.0973593498 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |