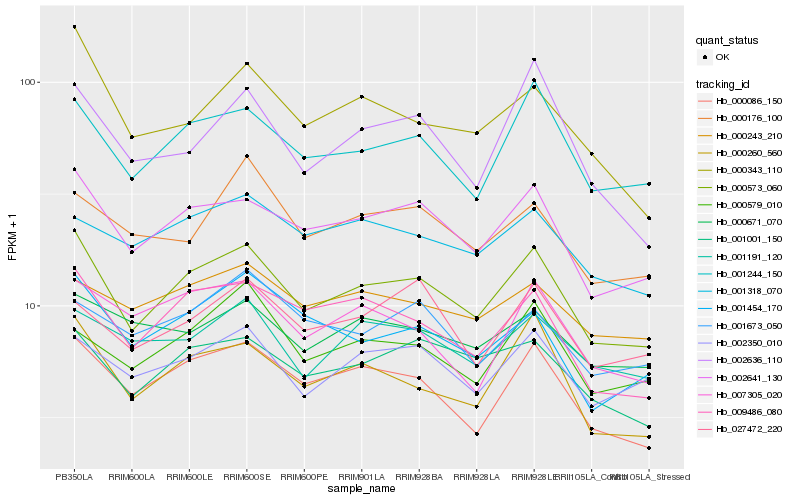
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000573_060 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 2 |
Hb_002641_130 |
0.0520884627 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 3 |
Hb_001244_150 |
0.0649166193 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 4 |
Hb_000086_150 |
0.0702733869 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000260_560 |
0.0741946102 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 6 |
Hb_009486_080 |
0.074221874 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 7 |
Hb_001001_150 |
0.0782372009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002350_010 |
0.0841641485 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 9 |
Hb_001454_170 |
0.0864070827 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 10 |
Hb_000343_110 |
0.0885773493 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 11 |
Hb_000579_010 |
0.0905255341 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 12 |
Hb_001191_120 |
0.0914811399 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002636_110 |
0.0935731256 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 14 |
Hb_007305_020 |
0.0942774327 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 15 |
Hb_001318_070 |
0.0943939916 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_027472_220 |
0.0955587182 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_000243_210 |
0.095577525 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000671_070 |
0.0964792899 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 19 |
Hb_000176_100 |
0.0967610695 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 20 |
Hb_001673_050 |
0.0985577055 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |