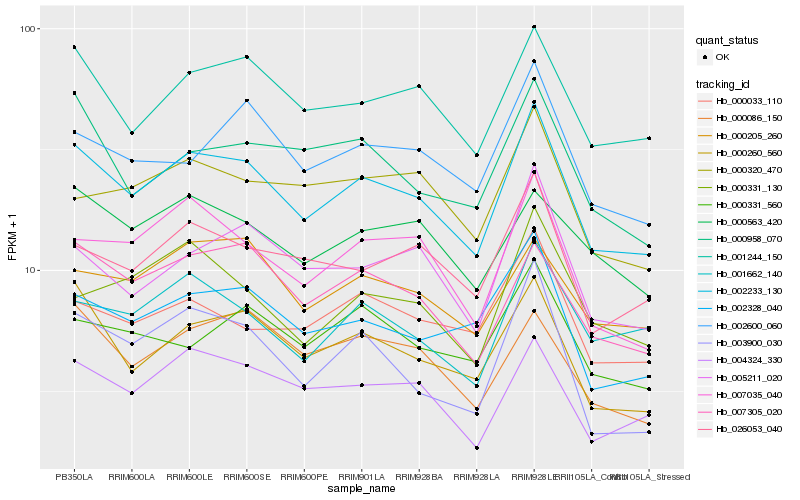
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002233_130 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_001244_150 |
0.0711161375 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 3 |
Hb_000260_560 |
0.0738080074 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004324_330 |
0.0743045751 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 5 |
Hb_007305_020 |
0.077962797 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 6 |
Hb_007035_040 |
0.0797985618 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002328_040 |
0.081917604 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 8 |
Hb_026053_040 |
0.0826659615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005211_020 |
0.086344189 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002600_060 |
0.0871031575 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 11 |
Hb_000205_260 |
0.0883785579 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 12 |
Hb_001662_140 |
0.0896234594 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000958_070 |
0.0899866258 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 14 |
Hb_000086_150 |
0.0902879064 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003900_030 |
0.0907729127 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000033_110 |
0.0924260897 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 17 |
Hb_000563_420 |
0.0932138948 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 18 |
Hb_000331_560 |
0.093666297 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 19 |
Hb_000320_470 |
0.0952822816 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 20 |
Hb_000331_130 |
0.0960598647 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |