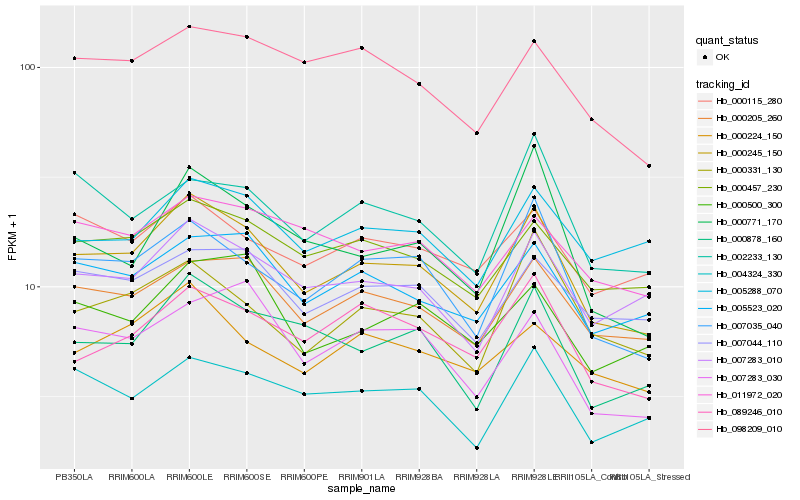
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000245_150 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 2 |
Hb_007035_040 |
0.075475015 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000205_260 |
0.0762292704 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 4 |
Hb_005523_020 |
0.0815129424 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 5 |
Hb_007283_010 |
0.0823058821 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 6 |
Hb_000457_230 |
0.0832879163 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000878_160 |
0.0862392131 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 8 |
Hb_007044_110 |
0.0905044692 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000224_150 |
0.0920737146 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004324_330 |
0.0933006406 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 11 |
Hb_089246_010 |
0.0947447082 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 12 |
Hb_000500_300 |
0.0951831088 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 13 |
Hb_098209_010 |
0.096304361 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 14 |
Hb_000331_130 |
0.0969540781 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005288_070 |
0.0974831401 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000115_280 |
0.0981349765 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_011972_020 |
0.0986396434 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_007283_030 |
0.0991087321 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 19 |
Hb_002233_130 |
0.0996212918 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_000771_170 |
0.0997100335 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |