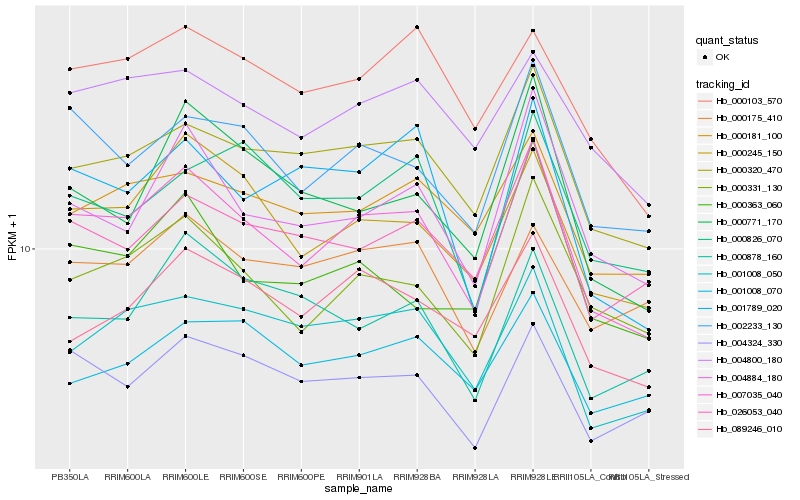
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007035_040 |
0.0 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000245_150 |
0.075475015 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 3 |
Hb_000320_470 |
0.0766182492 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_002233_130 |
0.0797985618 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_000331_130 |
0.0802305248 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004884_180 |
0.0871984556 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001008_050 |
0.0897141408 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 8 |
Hb_026053_040 |
0.0929427015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004324_330 |
0.0934677118 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 10 |
Hb_000181_100 |
0.0950990689 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 11 |
Hb_089246_010 |
0.0954363538 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 12 |
Hb_004800_180 |
0.0956644531 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 13 |
Hb_000103_570 |
0.0978263717 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000771_170 |
0.1017390813 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_001789_020 |
0.1030086584 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000175_410 |
0.1072619378 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000878_160 |
0.1075975779 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 18 |
Hb_000826_070 |
0.1086293274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001008_070 |
0.1089173116 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 20 |
Hb_000363_060 |
0.1090260664 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |